Strong Association of Polymorphism in SPRED2 Gene with Disease Susceptibility and Clinical Characteristics of Rheumatoid Arthritis in the Iranian Population
-
Pakzad, Bahram
-
Division of Rheumatology, Department of Internal Medicine, School of Medicine, Isfahan University of Medical Sciences, Isfahan, Iran
-
Moghadammanesh, Hamed
-
Division of Rheumatology, Department of Internal Medicine, School of Medicine, Isfahan University of Medical Sciences, Isfahan, Iran
-
Salesi, Mansour
-
Division of Rheumatology, Department of Internal Medicine, School of Medicine, Isfahan University of Medical Sciences, Isfahan, Iran
-
 Salehi, Rasoul
Department of Genetics and Molecular Biology, School of Medicine, Isfahan University of Medical Sciences, Isfahan, Iran, rasoulsalehii@yahoo.com
Salehi, Rasoul
Department of Genetics and Molecular Biology, School of Medicine, Isfahan University of Medical Sciences, Isfahan, Iran, rasoulsalehii@yahoo.com
-
Department of Genetics and Molecular Biology, School of Medicine, Isfahan University of Medical Sciences, Isfahan, Iran
Abstract: Background: The high heritability of Rheumatoid Arthritis (RA) has been estimated from different studies. Recently, Genome-Wide Association Studies (GWAS) show a large number of Single Nucleotide Polymorphisms (SNPs) loci affecting susceptibility to RA. The rs934734 polymorphism in the SPRED2 gene is one of these loci. Studies have shown that the SPRED2 gene is involved in the regulation of inflammatory response, leukocyte infiltration, and local chemokine production. In the current study, the possible association between SNP rs934734 (intronic variant) in the SPRED2 gene with RA risk in the Iranian population was evaluated.
Methods: One hundred fourteen RA patients and 120 healthy counterparts were recruited in this case-control study to evaluate rs934734 genotypes using the real-time PCR High Resolution Melting method (HRM).
Results: Logistic regression analysis demonstrated that GG and AG genotypes compared with AA genotype increase the risk of RA (GG vs. AA; OR=4.61; 95%CI [2.21-9.35]; p<0.001 and AG vs. AA; OR=2.54; 95%CI [1.36-4.76]; p=0.004). Furthermore, subjects with allele G were more frequently affected with RA than subjects with A allele (OR=2.33; 95%CI [1.61-3.38]; p<0.001). Besides, in the patient group, there was a significant correlation between Erythrocyte Sedimentation Rate (ESR) and C-reactive protein (CRP) concentration with rs934734 polymorphism (p<0.05).
Conclusion: Our findings suggest that rs934734 in SPRED2 strongly underlies RA development and is associated with clinicopathological characteristics of this disease.
Introduction :
Rheumatoid Arthritis (RA), classified as an autoimmune disease, is a long-term progressive disorder that causes inflammation and painful swelling in and around the joints and other body organs 1,2. The prevalence of RA varies between 0.5 and 1% and begins at any age, but its incidence increases with age 3,4. RA, similar to most of the other autoimmune diseases, displays a striking imbalance between the two sexes, with two or three times higher prevalence in women than men 5,6.
RA has a multifactorial etiology as the interaction of several genetic and environmental factors is essential for the development of this disorder 7. Multiple studies have shown that genetic factors are critically involved in the incidence of RA 8. With regard to twin studies, the heritability of RA has been estimated to be 50-60% 9. Furthermore, some works demonstrated that a positive family history of RA increases the risk of this disease 3-9 folds 10.
The most frequent type of variation in the human genome is Single Nucleotide Polymorphism (SNP) with prevalence of one in every 300 nucleotides which could be associated with diseases such as RA 11-13. With recent advances in genotyping and sequencing techniques, studies have shown a large number of SNPs loci that impact the susceptibility to RA. For instance, more than 100 risk-bearing loci for RA have been identified in people of European and Asian descent by Genome-Wide Association Studies (GWAS) 14,15. Several GWAS that have been conducted to recognize new predisposing genes of the RA disease have revealed that the rs934734 in sprouty-related protein with EVH1 domain 2 (SPRED2) gene is one of the candidate loci 16.
SPRED2 gene (located in chromosome 2) is a member of the SPRED family of proteins and negatively regulates the Ras/Raf/ERK/MAPK pathway 17,18. Studies on Spred2 knockout (Spred2-/-) mice demonstrated that this gene negatively regulates inflammatory response in the colonic mucosa, leukocyte infiltration, and local chemokine production in polymicrobial septic peritonitis 19-21. The GWAS also demonstrated that Spred2 gene is a candidate gene for the development of IBD 22.
The rs934734 (g.65595586G>A) is an intronic variant of the SPRED2 gene. GWAS demonstrated that the G allele of rs934734, as a risk factor, increases the risk of RA development 23,24. In the present study, for the first time, the possible association between SNP rs934734 in the SPRED2 gene with RA risk in the Iranian population was assessed. Also, the possible influence of association between some demographic and laboratory characteristics and rs934734 genotypes on susceptibility to RA was assessed.
Materials and Methods :
Study population and sample preparation
In this case-control study, a total of 114 unrelated subjects with RA as a case group (mean age: 47.39± 10.50 years) and 120 unrelated healthy subjects as a control group (mean age: 45.39±12.73 years) were included. Patients were recruited from Alzahra Hospital in Isfahan city of Iran. All the RA patients met the diagnostic criteria introduced by the American College of Rheumatology (ACR). All healthy controls and their families had no symptoms or history of RA or other immunological and autoimmune disorders. The participants of the study were interviewed and data on sex, age (at sampling time and onset), Body Mass Index (BMI, calculated as weight [kg] divided by height [m2] squared), blood pressure, and family history of RA and other autoimmune conditions were obtained using a structured questionnaire. Also, laboratory characteristics such as Erythrocyte Sedimentation Rate (ESR), C-reactive protein (CRP), White Blood Cell (WBC) count, hemoglobin, Platelet count test (PLT), creatinine, Blood Urea Nitrogen (BUN), Fasting Blood Sugar (FBS), High-Density Lipoprotein (HDL), Low-Den-sity Lipoprotein (LDL), and Triglyceride (TG) were recorded. This study was approved by the university ethics board (Approved number: IR.MUI.REC.1397. 102) and all participants gave written informed consent.
DNA extraction and genotyping of polymorphism
Approximately, 5 ml of venous blood was collected into EDTA anticoagulant tubes from each participant and stored at −20°C for DNA isolation. Genomic DNA was extracted from 200 μl of peripheral blood samples using a kit (GenetBio Co., Korea) consistent with the instruction manual. The purity and concentration of all genomic DNA samples were assessed by agarose gel electrophoresis and spectroscopy at wavelengths of 260 and 280 nm, respectively, and then DNA was stored at −20°C.
The real‑time polymerase chain reaction high‑reso-lution melting (HRM) method was used to determine rs934734 polymorphism genotypes. HRM was performed using Type-it HRM PCR kit that contained HotStarTaq plus DNA polymerase and EvaGreen dye (Qiagen GmbH, Germany), and the analysis was carried out with Rotor-Gene 6000™ (Corbett Research Pty Limited, Australia). The forward and reverse primer sequences for the 180-bp fragment that spanned the rs934734 in the SPRED2 gene were CCTCCTCA CGAGGACTGC and TGAACTGTGAACTTATTTCT CCAAA, respectively. The thermal profile of the reaction was 5 min of denaturation at 95°C, 40 cycles of 95°C for 10 s, 58°C for 30 s and 72°C for 20 s. The melting curve was generated by increasing the temperature between 65°C and 95°C at the rate of 0.1°C/s. Finally, melting curves were normalized among the two temperatures to determine the specimens with known genotypes as standard. For utilizing sample genotypes in HRM analysis as a standard, some samples were subjected to direct Sanger sequencing and their correct genotypes were determined.
Statistical analyses
The SPSS software v22 (IBM, USA) was used for statistical analyses. The allele and genotype frequencies were tested for Hardy Weinberg equilibrium by the χ2 test. Logistic regression analysis was accomplished to investigate the association between genotypes and RA and to calculate specific Odds Ratios (ORs), 95% Confidential Intervals (CIs), and p-values. For demographic, clinical, and laboratory characteristics, p-values were calculated using independent sample t‑test, Chi‑square or Mann–Whitney U test. The significance level was set at p<0.05.
Results :
To evaluate the correlation between rs934734 polymorphism with RA incidence, totally 234 subjects in case and control groups were analyzed among whom 114 patients (82 females and 32 males, mean age at sampling time: 47.39±10.50) were in the case group and 120 (81 females and 39 males, mean age at sampling time: 45.39±12.73) were healthy subjects in the control group. Demographic and clinical characteristics of subjects who were included in the study are shown in table 1. There was no substantial correlation between case and control groups in terms of age (p= 0.189) and gender (p=0.480), demonstrating that for these variables matching was adequate. From all patients, twenty (17.5%) patients had a positive family history whereas controls did not have a history of RA or other autoimmune diseases. Based on laboratory tests, ESR, CRP, WBC count, and creatinine were significantly higher in patients than in healthy controls (p<0.05) while hemoglobin level in control group was significantly higher in comparison to patients group (p<0.001). Positive RF was observed in all patients. Other laboratory factors including PLT, BUN, FBS, HDL, LDL, and TG were not significantly different be-tween patients and healthy controls (p>0.05). The details of laboratory characteristics of patients with RA and healthy controls are presented in table 2.
Genotype and allele distribution
In this study, it was demonstrated that the genotype distribution of rs934734 polymorphism in two groups was in agreement with Hardy-Weinberg equilibrium. Significant association was found between GG (compared with AA; p<0.001) and AG (compared with AA; p=0.004) genotypes with the risk of RA. The frequency of AA, AG, and GG genotypes was 20.2%, 46.5%, and 33.3% in patients and 44.2%, 40%, and 15.8% in controls, respectively (Table 3). Analysis of the combined genotype demonstrated that the GG+AG increases the risk of RA compared with the AA genotype (p<0.001). In allele distribution analysis, an increased level of allele G in patients (56.6%) was observed when compared with controls (35.8%), which was statistically significant (p<0.001). Furthermore, stratification based on sex and age of onset displayed a non-significant correlation between these subgroups and SNP genotypes in patients (p=0.183 and p=0.870, respectively). The mean concentration of ESR and CRP in the patient group was significantly different in genotype stratification (p<0.05) while there was no significant association between BMI, creatinine, and hemoglobin with this polymorphism (p>0.05) (Table 4).
Discussion :
Convergent lines of evidence indicated that a large number of SNPs loci affect susceptibility to autoimmune disorders 25,26. Several GWAS demonstrated that numerous SNPs loci in different genes are associated with RA 15,27. The association of rs934734 polymorphism in the SPRED2 gene (located at chromosome 2p14) with RA was revealed by two GWAS 23,24. Researches have shown that the SPRED2 gene is involved in the regulation of inflammatory response, leukocyte infiltration, and local chemokine production 19,20. Therefore, dysregulation of this gene could be involved in autoimmune pathogenesis. To the best of our know-ledge, the present study was the first single locus association study that showed the association between rs934734 polymorphism in SPRED2 gene with the risk of RA. In our study, logistic regression analysis de-monstrated that homozygous GG and heterozygous AG genotypes compared with the AA genotype increase the risk of RA (GG vs. AA; OR=4.61; 95%CI [2.21-9.35] and AG vs. AA; OR=2.54; 95%CI [1.36, 4.76]). Likewise, combined genotype analyses indicated that GG+AG compared with the AA genotype increases the risk of disease (OR=3.13; 95%CI [1.32-5.62]). Moreover, individuals with allele G were more frequently affected with RA than subjects with A allele (OR= 2.33; 95%CI [1.61, 3.38]) (Table 3). Our analysis was consistent with a GWAS that carried out on people of European descent and revealed that the G allele is a risk factor for RA 19. However, these results were not confirmed in the other GWAS on African-American populations 28. Furthermore, Mizuno et al reported that this polymorphism was not correlated with the progression of functional disability in RA patients 29.
Besides, in the patient group, a significant correlation between the sex group, the mean age of onset, and also ESR and CRP concentration and different genotypes of rs934734 polymorphism (p<0.05) was found (Table 4). One of the main limitations of the current study was the small sample size.
Conclusion :
Our results showed that rs934734 in SPRED2 is a strong determinant for the development of RA and clinicopathological characteristics of this disease. In our previous studies, similar results were also found about association of specific polymorphisms with clini-co-pathological characteristics of RA disease including variants in NLRP3, NOD2, and microRNA-124 genes 30-32.
However, performing replication studies in different populations is a necessity to validate these results. Finally, further association studies with larger sample size would help to confirm the suggested correlations. Also, other polymorphisms that were not included in our study might be involved in determining the risk of RA, thus future studies are necessary.
Acknowledgement :
We would like to appreciate any support provided by Isfahan University of Medical Sciences.
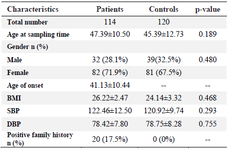
Table 1. Baseline characteristics of RA patients and control subjects
Data are mean±SD, or n (%). * p-value<0.05; SD: Standard deviation; RA: Rheumatoid arthritis; BMI: Body mass index; SBP: Systolic blood pressure; DBP: Diastolic blood pressure.
|
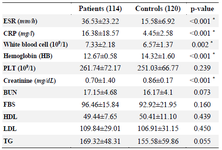
Table 2. Laboratory characteristics of RA patients and control subjects
Data are mean±SD, or n (%). * p<0.05; SD: Standard deviation; RA: Rheumatoid arthritis; ESR: Erythrocyte sedimentation rate; CRP: C‑reactive protein; BUN: Blood urea nitrogen; PLT: Platelet; HDL: High‑density lipoprotein; LDL: Low‑density lipoprotein; TG: Triglyceride; FBS: Fasting blood sugar.
|
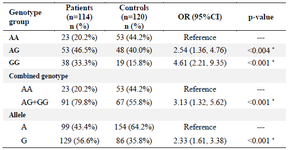
Table 3. Association between genotypes and allele frequency with RA risk
* p-value<0.05.
|
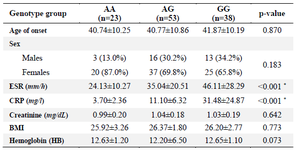
Table 4. Stratification analysis of the SPRED2 polymorphism (rs934734) in patients
Data are mean±SD, or n (%). * p-value<0.05. ESR: Erythrocyte sedimentation rate; CRP:C‑ reactive protein; BMI: Body mass index; SD: Standard deviation.
|
|