Comparative Expression of ADAMTS2, CA1, and OLAH in Immune Cells of Rheumatoid Arthritis Patients: Real-time PCR Study on In-silico analysis of Treated vs. Newly Diagnosed Patients
-
Dehghan, Fatemeh
-
Laboratory Sciences Research Center, Golestan University of Medical Sciences, Gorgan, Iran
-
Bazi, Zahra
-
Department of Medical Biotechnology, Faculty of Advanced Medical Technologies, Golestan University of Medical Sciences, Gorgan, Iran
-
Cancer Research Center, Golestan University of Medical Sciences, Gorgan, Iran
-
Aghaei , Mehrdad
-
Golestan Rheumatology Research Center, Golestan University of Medical Science, Gorgan, Iran
-
Gholizadeh, Masoomeh
-
Metabolic Disorders Research Center, Biomedical Research Institute, Golestan University of Medical Sciences, Gorgan, Iran
-
Malakouti, Romina
-
Laboratory Sciences Research Center, Golestan University of Medical Sciences, Gorgan, Iran
-
 Hesari, Zahra
Department of Biochemistry, Faculty of Medicine, Golestan University of Medical Sciences, Gorgan, Iran, Tel: +98 9120766506, Fax: +98 17 32456242; E-mail: Hesari7@yahoo.com
Hesari, Zahra
Department of Biochemistry, Faculty of Medicine, Golestan University of Medical Sciences, Gorgan, Iran, Tel: +98 9120766506, Fax: +98 17 32456242; E-mail: Hesari7@yahoo.com
Abstract: Background: Rheumatoid Arthritis (RA) is a chronic inflammatory joint disease. Current treatments often have limited efficacy and cause side effects due to their nonspecific action, while early diagnosis is challenging. This study combined bioinformatics and experimental methods to identify key genes and pathways involved in RA, aiming to discover novel therapeutic targets and diagnostic biomarkers.
Methods: RNA-seq data from immune cells of RA patients and healthy donors (GSE117769) were analyzed with DESeq to identify Differentially Expressed Genes (DEGs). Affected pathways were explored using EnrichR, and druggable genes were identified through DGIdb and a literature review. Expression of candidate genes was validated in additional RA blood and synovium microarray datasets (GSE45291, GSE82107, GSE77298) using the GEO2R tool. Finally, RT-qPCR was used to measure the expression of selected genes in Peripheral blood Mononuclear Cells (PBMCs) from newly-diagnosed and chronic RA patients and controls, with associations to clinical features and diagnostic accuracy assessed. Synovial fluid of RA patients were stained with Giemsa.
Results: Combined in-silico and experimental analysis demonstrated significant upregulation of CA1, OLAH, and ADAMTS2 in the PBMCs of RA patients. However, only ADAMTS2 showed high expression in the synovial tissue of these patients. While OLAH and ADAMTS2 were predominantly overexpressed in newly-diagnosed cases, CA1 levels were consistently elevated in both early and chronic stages of RA.
Conclusion: This study identified CA1, OLAH, and ADAMTS2 as being upregulated in RA, with ADAMTS2 showing promise as a therapeutic target, suggesting it may also have potential as a candidate for diagnosis and treatment.
Introduction :
Prolonged inflammation of the synovial joints is a hallmark of Rheumatoid Arthritis (RA), a chronic, progressive autoimmune disease characterized by persistent joint inflammation and swelling. Between 0.5 and 1% of the global population is affected by this disease 1. This illness is characterized by joint tenderness, pain, and swelling 1, which hinders diarthrodial joints' normal function and leads to severe physical impairment, mental anguish, and financial hardship 2,3. The effective management of RA requires a comprehensive, multidisciplinary approach that focuses on symptom control, joint preservation, and improving overall patient quality of life. The primary pharmacological strategy involves the administration of Disease-Modifying Antirheumatic Drugs (DMARDs), often in combination with adjunctive therapies. DMARDs function to attenuate disease progression and maintain joint integrity and function. These agents are broadly categorized into two main classes: nonbiologic DMARDs (e.g., methotrexate, sulfasalazine) and biologic DMARDs (e.g., tumor necrosis factor inhibitors, interleukin-6 receptor antagonists). Adjunctive pharmacological agents, employed as supportive therapies, commonly include corticosteroids, Nonsteroidal Anti-Inflammatory Drugs (NSAIDs), immunosuppressive agents, and analgesics. Although these therapeutic interventions are widely utilized, they may demonstrate limited efficacy in specific patient subsets. Furthermore, prolonged use is associated with clinically significant adverse effects, including but not limited to hypertension, dyslipidemia, and insulin resistance 4-6.
The discovery of new gene networks and molecular pathways associated with the etiology of RA holds significant potential for the development of targeted treatment approaches. Recent developments in high-throughput technologies, specifically RNA sequencing (RNA-seq) and microarray analysis, have significantly improved functional genomics research and gene discovery. The removal of probe-based detection limitations, increased sensitivity, a wider dynamic range, and the ability to detect transcripts that have not yet been annotated are just a few of the benefits that RNA-seq has over microarrays, in particular. The availability of transcriptomic data generated by these techniques is increasing, thanks to open repositories such as the Gene Expression Omnibus (GEO). These datasets, when paired with advanced bioinformatics techniques, allow for extensive, integrative analyses that allow for the methodical discovery of new biomarkers and possible treatment targets for RA and other multifactorial 7.
The goal of this study was to identify differentially expressed genes associated with the pathophysiology of RA to identify potential therapeutic genes for the disease by analyzing RNA sequencing data from leukocyte samples downloaded from the GEO database. By conducting comprehensive bioinformatics analysis and verifying ADAMTS2, CA1, and OLAH genes using Real-Time PCR in samples from people with early-stage and chronic RA, the study seeks to discover new biomarkers and molecular targets.
Carbonic anhydrase 1 (CA1) is a zinc metalloenzyme belonging to a large family of carbonic anhydrase isoforms, which catalyze the reversible hydration of carbon dioxide (CO₂) to bicarbonate (HCO₃⁻) and protons (H⁺) 8. CA1, may influence the migration of Fibroblast-Like Synoviocytes (FLS) within inflamed joints in RA, thus contributing to disease progression 9. Supporting this, transgenic mouse models overexpressing CA genes exhibit exacerbated RA symptoms and enhanced joint destruction 10.
ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) is a family of multidomain extracellular protease enzymes. It has been shown that the inflammatory cytokine IL-1α—known to be pivotal in cartilage and bone homeostasis and pathology—can upregulate ADAMTS-2 and ADAMTS-3 expression in osteoblast-like cell lines. This raises the possibility that pro-inflammatory cytokines such as IL-1α may similarly induce ADAMTS2 expression in RA, contributing to joint degradation 11.
Oleoyl-ACP hydrolase (OLAH) has critical role in fatty acid biosynthesis. Fatty acids are increasingly recognized for their modulatory effects on immune responses, influencing inflammation in diverse ways. For example, short-chain fatty acids have been shown to suppress the production of pro-inflammatory cytokines such as IL-1β, IL-6, and TNFα, thereby exerting anti-inflammatory effects. In contrast, medium-chain fatty acids can promote inflammation through activation of Toll-Like Receptor 2 (TLR2) 12.
Materials and Methods :
Identification of druggable targets in RA via integrated bioinformatics: Following a comprehensive analysis of the GEO database, dataset GSE117769 was selected, which includes RNA-seq data from white blood cells of 50 RA patients and 50 matched healthy controls, balanced for age, gender, and ethnicity. The dataset encompasses expression profiles of 36,765 genes. Normalization of the count data was conducted using the DESeq2 package, which models gene expression counts via a negative binomial distribution and accounts for library size differences. Differentially Expressed Genes (DEGs) were identified based on cut-off thresholds of log fold change (logFC) >2 and adjusted p<0.05, with p-values corrected using the Benjamini-Hochberg False Discovery Rate (FDR) method. Enrichment analysis of associated biological pathways was performed using the EnrichR database. To identify potential druggable targets, genes were analyzed via the Drug-Gene Interaction Database (DGIdb 3.0) 13. Molecular functions and biological processes of candidate genes were further explored using the GeneCards database. Finally, validation of candidate gene expression was performed using the GSE45291 dataset, which contains whole blood transcriptomic profiles from 513 RA patients and 20 healthy controls obtained via microarray analysis. Differential gene expression analysis was performed using GEO2R, which applies the Robust Multi-array Average (RMA) method for background correction and normalization 14.
Validation of target gene expression in RA synovial tissue using GEO data: To investigate gene expression profiles of candidate genes within the synovial tissue microenvironment, two publicly available Gene Expression Omnibus (GEO) datasets: GSE82107 and GSE77298 were analyzed. Dataset GSE82107 comprises synovial biopsy samples from 10 Osteoarthritis (OA) patients and 7 healthy controls, while GSE77298 includes samples from 16 RA patients and 7 healthy controls. Both datasets were generated using the Affymetrix Human Genome U133 Plus 2.0 Array platform 15. Differential gene expression analysis was performed using GEO2R, significantly dysregulated genes were selected with an adjusted p<0.05 using the Benjamini-Hochberg method. Enrichment analysis of associated biological pathways was conducted using the EnrichR database to gain insight into the functional implications of the identified genes.
Participant selection and grouping in RA study: Blood samples were collected from 60 male and female individuals aged 30-60 diagnosed with RA according to the American College of Rheumatology 2010 criteria (ACR 2010). The patients were divided into two groups: Group 1 included 20 newly diagnosed, treatment-naïve RA patients who had not yet received medications such as hydroxychloroquine, methotrexate, or corticosteroids, while Group 2 consisted of 20 chronic RA patients who had been undergoing treatment with these medications for at least six months. Additionally, a control group (Group 3) of 20 healthy individuals without any history of inflammatory or autoimmune diseases in the past three months was included. Exclusion criteria for the study ruled out participants with other autoimmune or inflammatory diseases, alcohol use, pregnancy or recent childbirth, and any malignancies or cancers.
Collecting samples and gene expression assay using qRT-PCR: Collecting samples and gene expression assay using qRT-PCR. Ten ml of fresh peripheral blood was collected in K3 EDTA tubes to prevent clotting. Peripheral Blood Mononuclear Cells (PBMCs) contain several types of cells such as lymphocytes, monocytes and macrophages. It represents the only site of active gene expression in the blood. PBMCs were isolated using Ficoll density gradient centrifugation. Total RNA was extracted from PBMCs using RNXplus reagent following the manufacturer’s instructions. RNA quality and quantity were assessed by measuring the absorbance ratio (A260/A280) with a spectrophotometer and integrity was confirmed by agarose gel electrophoresis. cDNA synthesis was performed using 1 µg of total RNA and the CinnaGen first-strand cDNA synthesis kit according to the manufacturer’s protocol. Negative control reactions without reverse transcriptase (NRT) were included to monitor for genomic DNA contamination.
Gene expression of target genes was measured by real-time PCR on the ABI StepOne Plus system using SYBR Green Master Mix. GAPDH was used as the internal control and normalization strategy. Cycling conditions were as follows: initial denaturation at 95°C for 10 min followed by 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. Melt curve analysis verified amplification specificity. Experiments were conducted in triplicate with at least three biological replicates.
Design and preparation of primers: The sequences of the target genes were retrieved from the NCBI nucleotide database (https://www.ncbi. nlm.nih.gov/nucleotide). Primers were designed using Gene Runner 6.0 software. To ensure specificity for cDNA and avoid amplification of contaminating genomic DNA, primers were designed to span exon-exon junctions, a widely accepted strategy in qRT-PCR primer design. Primer melting temperature (Tm) and secondary structures were evaluated using the OligoAnalyzer tool (https://www.idtdna.com/pages/tools/oligo-analyzer)1. Primer sequences provided in (Table 1).
Synovial fluid analysis: Synovial fluid samples were collected sterilely from patients with newly diagnosed rheumatoid arthritis in tubes containing anticoagulant. The samples were examined morphologically under a microscope using Neubauer slides (hemocytometer) and stained with Giemsa stain.
Statistical analysis: Data were analyzed using SPSS 16 and GraphPad Prism 9 with one-way ANOVA, Shapiro-Wilk, Mann-Whitney, and Student’s t-tests. Results are presented as Mean±SD from at least two experimental replicates, with significance set at p<0.05.
Results :
ADAMTS2, CA1 and OLAH genes nominated as candidate targets for RA: Analysis of 36,765 genes revealed that 19 genes were significantly differentially expressed in the RA group, defined by a log fold change (LogFC) >2 and an adjusted p<0.05. However, pathway enrichment analysis of these differentially expressed genes did not yield any statistically significant pathways (Table 2). Subsequent bioinformatics analysis identified three of the upregulated genes as druggable targets (Table 3). To further validate their relevance as potential biomarkers or therapeutic targets, the expression of these genes was assessed in a larger independent microarray dataset. All three candidate genes were significantly upregulated in this dataset as well (Tables 3 and 4).
Transcriptomic evidence highlights ADAMTS2 upregulation in arthritic synovial tissue: Gene expression in synovial tissue was analyzed using GEO datasets, specifically the GSE77298 and GSE82107 files. In the GSE77298 dataset, 187 genes were significantly upregulated (fold change >2), while 471 genes were downregulated (fold change <-2). In the GSE82107 dataset, 55 genes were significantly upregulated (fold change >2), and 478 genes were downregulated (fold change <-2). Notably, the IL-17 signaling pathway was the main common pathway among the upregulated genes in both datasets, whereas pathways related to cardiac function and heart muscle disease were commonly and significantly enriched among the downregulated genes (Tables 5 and 6). According to both datasets, ADAMTS2 was significantly overexpressed in synovial biopsies from osteoarthritis and RA patients, while OLAH expression was decreased in both conditions. In contrast, the expression of the CA1 gene showed variability; interestingly, among the different isoforms of the CA gene, CA12 was consistently increased in both tissues (Table 7).
Participant characteristics: The participants included in this investigation were categorized into three distinct groups based on their age range of 30-60 years. It should be noted that the allocation of individuals in each group was done in a way that ensured the matching of both age and sex across all groups. The mean age of the participants in groups 1 to 3 was found to be 49±6.17, 49±6.44, and 49±6.97 years, respectively. This observation was made with a p-value of 0.882. Furthermore, it is worth mentioning that among the total participants, 7 individuals (35%) were males and 13 individuals (65%) were females, with this distribution being consistent across all three groups.
Elevated CA1, ADAMTS2, and OLAH expression in peripheral blood of RA patient: Figure 1 shows that all target genes were overexpressed in PBMCs of RA patients, with varying levels. The CA1 gene increased over 5-fold in both new and chronic groups compared to controls (p<0.038 and p<0.002). ADAMTS2 expression was also significantly elevated, especially in new cases with a 2-fold higher increase than chronic patients. In contrast, OLAH expression was notably high (over 12-fold) mainly in new RA cases.
Synovial fluid analysis: In image prepared from synovial fluid sample taken from the newly diagnosed RA patient showed an increase in leukocytes, especially neutrophils in the synovial fluid indicating an active inflammatory condition in the joints of these patients (Figure 2).
Discussion :
The development of RA involves complex molecular pathways that impair the immune system, leading to persistent inflammation. Joint damage is caused by immune cell migration into the synovium and the proliferation of synovial fibroblasts. The specific molecular mechanisms behind the onset and progression of RA are still poorly understood, although inflammation and immune complexes are acknowledged as major contributors to the disease. Because of this knowledge gap, current therapies are less effective and frequently fail to prevent joint damage 16. In the present study, bioinformatics analyses with Real-Time PCR experiments were combined to identify novel target genes for RA management. Integrated in silico and experimental data revealed significant upregulation of the enzymes CA1, OLAH, and ADAMTS2 in PBMCs of RA patients. Notably, only ADAMTS2 and another isoform of Carbonic anhydrase gene, CA12 exhibited high expression in synovial tissue. The ADAMTS family, characterized by disintegrin, metalloproteinase, and thrombospondin motifs, plays a crucial role in remodeling the extracellular matrix through the degradation of fibrillar collagens 17,18. While several members of this family, including ADAMTS-4, ADAMTS-5, ADAMTS-7, and ADAMTS-12, have been implicated in the pathogenesis of RA 19, the specific role of ADAMTS2 in RA remains poorly defined. Findings demonstrate that ADAMTS2 expression is significantly elevated in both the peripheral blood and synovial tissue of RA patients, suggesting its involvement in disease processes. Supporting this, prior research by Alper et al revealed that the inflammatory cytokine IL-1α—known to be pivotal in cartilage and bone homeostasis and pathology—can upregulate ADAMTS-2 and ADAMTS-3 expression in osteoblast-like cell lines 11. This raises the possibility that pro-inflammatory cytokines such as IL-1α may similarly induce ADAMTS2 expression in RA, contributing to joint degradation. Therefore, targeting ADAMTS2 may represent a promising therapeutic strategy to mitigate extracellular matrix breakdown and prevent joint damage in RA patients.
Carbonic anhydrase 1 (CA1) is a zinc metalloenzyme belonging to a large family of carbonic anhydrase isoforms, which catalyze the reversible hydration of carbon dioxide (CO₂) to bicarbonate (HCO₃⁻) and protons (H⁺) 8. This reaction is fundamental for maintaining pH homeostasis, a critical aspect of physiological equilibrium 9. Data demonstrate that CA1 is predominantly expressed in the blood cells of RA patients, whereas other isoforms, such as CA12, show increased expression in synovial tissue. Although both CA1 and CA12 catalyze the same biochemical reaction, they differ significantly in cellular localization and function; CA1 primarily regulates systemic acid-base balance in erythrocytes 20, while CA12, a membrane-bound isozyme expressed at epithelial surfaces, modulates local pH and has been implicated in pathological processes including tumor microenvironment adaptation under hypoxic conditions 21,22. Emerging evidence suggests that carbonic anhydrases, including CA1, may influence the migration of FLS within inflamed joints in RA, thus contributing to disease progression 9.
Supporting this, transgenic mouse models overexpressing CA genes exhibit exacerbated RA symptoms and enhanced joint destruction 10. Moreover, the study by Strowitzki and colleagues highlighted the role of carbonic anhydrase in regulating intracellular pH in monocytes and macrophages, which in turn modulates inflammatory gene expression. Pharmacological inhibition of carbonic anhydrases disrupts this pH-sensitive inflammatory signaling and impairs CO₂ sensing in macrophages 23.
Notably, acetazolamide, a carbonic anhydrase inhibitor, has been shown to attenuate inflammation and hypoxic pulmonary hypertension through modulation of immune cell activity 24. Together, these findings, aligned with our results, implicate CA1 overexpression in PBMC as a contributing factor to inflammatory responses during both acute and chronic phases of RA, likely via modulation of immune cell acidity. Therefore, targeting CA1 with specific inhibitors presents a promising therapeutic avenue for mitigating inflammation and joint damage in RA.
Oleoyl-ACP hydrolase (OLAH) has emerged as a gene of interest in RA due to its critical role in fatty acid biosynthesis. Fatty acids are increasingly recognized for their modulatory effects on immune responses, influencing inflammation in diverse ways. For example, short-chain fatty acids have been shown to suppress the production of pro-inflammatory cytokines such as IL-1β, IL-6, and TNFα, thereby exerting anti-inflammatory effects. In contrast, medium-chain fatty acids can promote inflammation through activation of TLR2 12. Furthermore, long-chain fatty acids, including palmitic acid, have been implicated in driving pro-inflammatory signaling by engaging multiple Toll-like receptor complexes such as TLR2/TLR1, TLR2/TLR6, and TLR4 25. Given these complex interactions, recent research has increasingly focused on how variations in fatty acid concentration, metabolism, and composition contribute to RA pathogenesis 26. The involvement of OLAH in fatty acid metabolism thus underscores its potential significance in modulating inflammatory pathways within RA and warrants further investigation.
Given that OLAH and ADAMTS2 were predominantly overexpressed in newly diagnosed RA patients, while CA1 levels remained consistently elevated throughout both early and chronic stages, our findings suggest that CA1 and OLAH primarily drive the initiation and propagation of inflammation, whereas ADAMTS2 is mainly involved in tissue remodeling and joint destruction. Collectively, these genes hold promise as biomarkers for early diagnosis and as potential targets for novel therapeutic interventions in RA. Nonetheless, this study is limited by a relatively small sample size, lack of functional assays to establish causality, and a focus on PBMCs rather than comprehensive joint tissue analysis. Future investigations with larger cohorts and mechanistic studies are warranted to validate these targets and elucidate their roles in RA pathophysiology.
Conclusion :
This integrative analysis identified CA1, OLAH, and ADAMTS2 as key dysregulated genes in RA, with ADAMTS2 consistently upregulated in both blood and synovial tissue. Their distinct expression patterns highlight their roles in inflammation and joint damage, supporting their potential as diagnostic biomarkers and therapeutic targets. These findings advance molecular insights into RA and lay the groundwork for future translational applications.
Ethics approval: During collecting data, the researchers obtained informed consent from the participants after explaining the purpose and objective of the study. This study was approved by the Ethics Committee of Golestan University of Medical Sciences (IR.GOUMS.REC.1400.421). All procedures performed agreed with the principles of the Declaration of Helsinki and later amendments.
Acknowledgement :
This study was supported [grant number 112510] approved by the Ethics Committee of Golestan University of Medical Sciences (IR.GOUMS.REC.1400.421).
Funding: This study was supported by Golestan University of Medical Sciences [grant number 112510].
Conflict of Interest :
The authors declare that they have no conflict of interest.
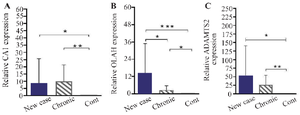
Figure 1. Results of changes in the expression of A) CA1 B) OLAH and C) ADAMTS2 genes in PBMC cells of new case rheumatoid arthritis patients, chronic patients, and control group.
|
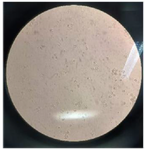
Figure 2. The image prepared from synovial fluid sample taken from newly diagnosed patients with rheumatoid arthritis. Leukocytes in a synovial fluid sample under Neubauer slide (Hemocytometer; mm3).
|
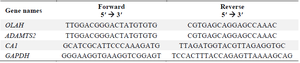
Table 1. The sequences of primers
|
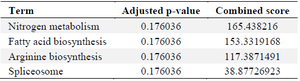
Table 2. Pathways that are regulated by upregulated genes (retrieved from EnrichR database)
|

Table 3. The properties and expression change of the selected genes in rheumatoid arthritis
|
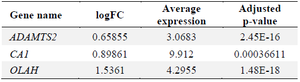
Table 4. Comparative analysis of ADAMTS2, CA1, and OLAH levels in microarray data
|
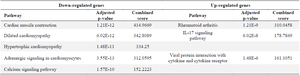
Table 5. Pathways affected by dysregulated genes in the synovial tissue of RA patients (GSE77298 dataset)
|
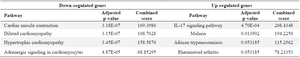
Table 6. Pathways influenced by dysregulated genes in the synovial tissue of RA patients (GSE82107 dataset)
|
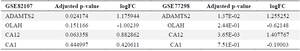
Table 7. Comparative analysis of ADAMTS2, CA1, and OLAH expression levels in the synovial tissue of RA patients
|
|