Optimization of RfxCas13d Expression in Escherichia coli Host using Response Surface Methodology
-
Abbaszadeh , Sepideh
-
Department of Medical Biotechnology, School of Advanced Medical Sciences and Technologies, Hamadan University of Medical Sciences, Hamadan, Iran
-
Research Center for Molecular Medicine, Institute of Cancer, Hamadan University of Medical Sciences, Hamadan, Iran
-
Eghbalsaied , Shahin
-
Research Center for Molecular Medicine, Institute of Cancer, Hamadan University of Medical Sciences, Hamadan, Iran
-
Soleimani, Meysam
-
Department of Pharmaceutical Biotechnology, School of Pharmacy, Hamadan University of Medical Sciences, Hamadan, Iran
-
Khazalpour, Sadegh
-
Department of Analytical Chemistry, Faculty of Chemistry and Petroleum Science, Bu-Ali Sina University, Hamadan, Iran
-
 Afshar , Saeid
Department of Medical Biotech-nology, School of Advanced Medical Sciences and Technolo-gies, Hamadan University of Medical Sciences, Hamadan, Iran, Tel: +98 81 38381796; Fax: +98 81 38380208; E-mail: safsharh@gmail.com
Afshar , Saeid
Department of Medical Biotech-nology, School of Advanced Medical Sciences and Technolo-gies, Hamadan University of Medical Sciences, Hamadan, Iran, Tel: +98 81 38381796; Fax: +98 81 38380208; E-mail: safsharh@gmail.com
-
Cancer Research Center, Institute of Cancer, Hamadan University of Medical Sciences, Hamadan, Iran
Abstract: Background: RfxCas13d, a key member of the Cas13 family, plays a vital role in CRISPR-based diagnostics for RNA sequence detection and gene silencing. This study aimed to enhance RfxCas13d expression by optimizing key parameters using Response Surface Methodology (RSM).
Methods: The plasmid pET28b-RfxCas13d-His (Addgene 141322) was introduced into BL21 (DE3) and Rosetta™ (DE3) strains. Initial expression tests were conducted, followed by RSM-guided optimization of factors such as isopropyl β-D-1-thiogalactopyranoside (IPTG) concentration, temperature, cell density at induction, and induction time in BL21 (DE3). Protein expression levels were quantified using ImageJ and AlphaEaseFC software to analyze band intensities.
Results: BL21 (DE3) was selected for further optimization based on preliminary results. Analysis of 26 RSM-designed experiments revealed that temperature, induction time, IPTG concentration, and their interactions significantly influenced RfxCas13d expression. Optimal conditions were identified as 0.25 mM IPTG, an OD600 nm of 0.8 at induction, 37°C, and Overnight (ON) of induction. The regression model exhibited high accuracy, with a correlation coefficient of 0.97 and a p-value less than 0.05, confirming a strong linear relationship between predicted and observed values.
Conclusion: This study highlights the significant impact of the four optimized factors on RfxCas13d expression. Under optimized conditions, a soluble protein concentration of 3.6 mg/100 ml cell culture was achieved after purification. It represents the first application of RSM for optimizing RfxCas13d expression, providing a foundation for further refinement of expression conditions. Continued use of RSM in future research will enhance the efficiency of RfxCas13d production for diagnostic and therapeutic applications.
Introduction :
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated (Cas) system was initially discovered as an adaptive immune mechanism in bacteria and archaea, enabling them to recognize and eliminate invading foreign genetic elements 1. In 2012, a groundbreaking discovery revealed that Cas9 protein, when combined with single-guide RNA (sgRNA), could precisely target and cleave specific DNA sequences in vitro 2, This finding established CRISPR-Cas9 as a revolutionary tool for gene editing 3,4. By 2016, the CRISPR-Cas9 system had been adapted for nucleic acid detection, further expanding its applications 5. The discovery of new CRISPR-Cas systems has considerably advanced nucleic acid recognition 6. Notably, the introduction of CRISPR-Cas12 and CRISPR-Cas13 into nucleic acid recognition frameworks has proven pivotal, leveraging their trans-cleaving mechanisms. These systems exhibit high detection accuracy and efficiency by identifying specific DNA and RNA targets, respectively, and triggering single-stranded DNA (ssDNA)/RNA trans-cleavage upon target recognition and cleavage 7,8. The Cas13 protein family is distinguished by two specific HEPN ribonuclease motifs that become active when they bind to target RNA, guided by gRNA 9. This family includes four known subtypes: Cas13a (C2c2), Cas13b, Cas13c, and Cas13d, with Cas13d being the smallest and most efficient for RNA targeting 10,11. Cas13-based systems have been widely employed in biosensor development, including the SHERLOCK technique and electrochemical biosensors, due to their high specificity and sensitivity 8,12,13. Among these, RfxCas13d distinguishes itself through its compact size, high specificity, and minimal off-target activity, establishing it as a leading tool for RNA targeting and editing. Its exceptional efficiency in mammalian cells further amplifies its potential for therapeutic and diagnostic applications. These attributes make RfxCas13d a highly adaptable and effective choice for RNA-based interventions, setting it apart from other Cas13 14,15. Given that numerous studies highlight the critical roles of oncogene transcription products and the overexpression of various non-coding RNAs, such as LncRNAs, in cancer development and progression, the distinct features of RfxCas13d make it a promising and versatile tool for driving future research advancements in this area 17,18.
For optimal recombinant protein production, it is crucial to optimize culture conditions like temperature, cell density at induction, and isopropyl β-D-1-thiogalactopyranoside (IPTG) concentration 19. The traditional One-Factor-at-a-Time (OFAT) method requires numerous experiments to find optimal conditions, but is often inefficient and fails to reveal interactions between variables 20. In contrast, the Design of Experiments (DoE) approach offers by identifying key factors and their interactions, leading to better predictions of optimal conditions with fewer trials 21. RSM within DoE has been successfully used to maximize recombinant protein yields by modeling the effects of various variables and their interactions 22,23. In RSM, two widely used experimental designs include the Box-Behnken Design (BBD) and the Central Composite Design (CCD) 24. Many studies have optimized both upstream and downstream processes using the RSM-Box-Behnken approach 25-28.
In this particular study, the RSM-Box-Behnken design was applied to optimize culture conditions for enhancing the total expression of Cas13. Four parameters- temperature, cell density at induction, IPTG concentration, and induction time-were analyzed for their individual and combined effects on protein expression.
Materials and Methods :
Bacterial cultivation was carried out using Luria-Bertani (LB) medium (Qlab, Canada). The Escherichia coli (E. coli) strains BL21(DE3) and Rosetta™(DE3) were obtained from the biobank of Hamadan University of Medical Sciences (Hamadan, Iran). For recombinant protein expression, the plasmid pET28b-RfxCas13d-His (Addgene plasmid #141322) was utilized. Antibiotics, including kanamycin and chloramphenicol, were purchased from Sigma-Aldrich (Germany). Additionally, a pre-stained protein marker (10-180 kDa, Cat. No. E-BC-R273) was acquired for protein analysis.
The bacterial strain and vector: pET28b-RfxCas13d-His system (Addgene 141322) includes the recombinant sequence of human RfxCas13d. This plasmid also features a kanamycin resistance gene and uses the T7 promoter, regulated by the lac promoter sequence, to drive the expression of RfxCas13d. The human-derived RfxCas13d sequence was expressed in both E. coli strains BL21 (DE3) and Rosetta™(DE3), which served as host cells harboring the recombinant plasmid for RfxCas13d protein production. The plasmid was successfully transformed into competent cells prepared using a cold calcium chloride solution, following the heat shock method as described by Sambrook et al 29. Selection of transformants occurred on LB agar plates supplemented with 50 µg/ml kanamycin for BL21 (DE3), 50 µg/ml kanamycin and 15 μg/ml chloramphenicol for Rosetta™(DE3), followed by cultivation in LB broth for 18 hr, and bacterial suspensions were stored in 25% (v/v) glycerol at -80°C for future use.
Initial expression analysis: For initial expression analysis, "Overnight (ON)" cultures of BL21 (DE3) and Rosetta™(DE3) containing the plasmid were diluted 1:100 into 5 ml of fresh culture media. These cultures were then incubated at 37°C while they were shaken until the Optical Density (OD) 600 reached 0.5. Subsequently, 1 ml of each culture was centrifuged, and the resulting cell pellets were stored at -20°C for later analysis. The remaining culture had IPTG (Sigma-Aldrich, Germany) added to a final concentration of 0.5 mM and was further incubated with shaking at 37°C for 3 hr and ON. For protein expression analysis, 1 ml samples from these cultures were centrifuged to collect the cells, which were then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a gel consisting of a 10% separating gel and a 5% stacking gel. Finally, The SDS-PAGE of separated proteins was visualized through staining with Coomassie Brilliant Blue.
Western blot: Western blot analysis involved the electro-transfer of proteins onto Amersham Protran 0.2 NC nitrocellulose Western blotting membranes at a setting of 30 mA for 14 hr, following the modified protocol provided by Sambrook et al 29. The membranes were blocked for 3 hr in Tris-Buffered Saline-Tween (TBST) containing 3% nonfat milk, washed three times with TBST, and incubated for 3 hr with Mouse Anti-His Tag HRP-conjugated Monoclonal Antibody (Catalog #MAB050H, 1:4000). The protein band was detected using an ECL kit (Catalog #:B111420, PARS Co. Mashhad, IR Iran) using FUSION FX chemiluminescence and fluorescence imaging.
Experiment designs: The factorial RSM-BBD methodology was employed to examine the effects of four independent variables including induction temperature, IPTG concentration, cell density at induction, and post-induction time (3 hr and ON), on RfxCas13d protein expression. The levels tested for each factor are presented in table 1. This analysis utilized Minitab 17 software to conduct 26 experimental runs. Statistical analysis through ANOVA 30 was applied to determine significant variables impacting protein expression. Expression levels of RfxCas13d in all experiments were quantified using high-quality images captured from SDS-PAGE gel evaluated with AlphaEaseFC and ImageJ software.
Purification of the soluble portion of recombinant rfxcas-13d: For purification, the soluble fraction of the recombinant rfxcas13d from a 100 ml culture under optimal conditions was resuspended in 25 ml of Lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10% v/v glycerol, 20 mM βME, pH=8.0) and maintained at 4°C. The cells were then sonicated for 20 min at 80% amplitude with intervals of 30 s on and 60 s off using bandelin sonopuls homogeniser while being maintained on ice. Following sonication, the lysate was centrifuged at 12,000 rpm for 25 min, and the pellet was removed and discarded. The supernatant was then purified using Ni-NTA agarose affinity chromatography under native conditions, following the manufacturer’s protocol (DNAbioteck, Iran). The column was washed with buffers containing 20 mM imidazole, and subsequently, the RfxCas13d protein was eluted using 250 mM imidazole. The quantification of the concentration of the purified protein was performed using the Bradford assay method and Bovine Serum Albumin (BSA) (Sigma-Aldrich) was utilized to prepare the standard curve 31.
Results :
Initial expression analysis: Initially, the recombinant protein's authenticity was confirmed through Western blot analysis, where the specific protein band's location was identified (Figure 1A). Furthermore, the protein expression was analyzed at two distinct intervals, 3 hr and ON, within BL21 (DE3) and Rosetta™ (DE3) strains. This assessment involved visual inspection of the protein bands on an electrophoresis gel, revealing no significant differences in protein expression between the two strains (Figure 1B). However, the presence of additional protein bands was significantly higher in the Rosetta™ (DE3) strain compared to the BL21 (DE3) strain, which could increase the complexity of downstream purification due to the presence of additional protein bands. Consequently, protein expression was fine-tuned specifically for the BL21 (DE3) strain.
Optimization of recombinant RfxCas13d expression: The RSM-BBD approach was employed to evaluate the effects of four variables- temperature of induction, IPTG concentration, cell density at induction, and post-induction time- on RfxCas13d expression. A four-level BBD incorporating 26 experimental runs was adopted. The expression levels of RfxCas13d across all trials were quantified through images obtained from SDS-PAGE gels and AlphaEaseFC software (Figure 2). The outcomes derived from these BBD experiments regarding RfxCas13d expression are summarized in table 1. Quadratic regression models for optimizing RfxCas13d expression were validated and confirmed through Analysis of Variance (ANOVA), with a p-value below 0.05 indicating statistical significance. ANOVA highlighted the model's significant impact on RfxCas13d expression (p-value<0.05) (Table 2). The normal probability plot illustrates the normal distribution of the residuals. In the Versus Fits Plot and the Versus Order Plot, the residuals exhibit random scatter, indicating that the model is well-fitted (Figures 3A-C). Validation tests corroborated that the experimentally obtained production values closely matched those predicted statistically, thereby verifying the model's accuracy. Figure 3D displayed a correlation of 0.97 and a p-value <0.05, suggesting a strong linear relationship between observed and predicted values.
Regarding the total protein production of RfxCas13d, ANOVA revealed significant model terms including Te, I, Ti, I2, Te*Ti, Te*I, O*I, O*Te, and O*Ti, while other terms were not significant due to p-values exceeding 0.05, leading to their exclusion (Table 2). Consequently, a simplified second-order polynomial equation was formulated for protein expression (response) based on significant factors, represented as I (IPTG concentration), Te (temperature), O (OD600 nm before induction), and Ti (post-induction time) (Equation).
Time and the interaction between OD600 nm before induction and time were identified as having the most substantial influence on the response. These were followed by temperature, the interaction of OD600 nm before induction with temperature, the square of IPTG concentration, as well as the interactions between temperature and time, IPTG concentration and OD600 nm before induction, and IPTG concentration (Figure 4). The interaction effects between these independent variables were further explored through three-dimensional response surface graphs (Figure 5).
The range of recombinant protein expression levels varied widely across the 26 designed experiments, from 1.75 to 7.27% turbidity as shown in table 1. Following the identification of optimal conditions (IPTG concentration=250 µM, temperature=37°C, OD600 nm before induction=0.8, and post-induction time at ON), recombinant RfxCas13d protein expression was executed and subsequently characterized by SDS-PAGE, as depicted in figure 6A. This characterization aimed to assess the model's suitability. The experimental outcome was then compared with data from the optimum point and predictive protein levels. The numerical equivalent of the predicted turbidity and the experimental result were calculated with Image J and AlphaEaseFC software, which were 8.45 and 8.2, respectively.
Protein purification: Initial tests on the expression of RfxCas13d recombinant protein under optimal conditions indicated that half of the protein was found in the soluble phase. The recombinant RfxCas13d protein was then successfully fractioned using a Ni-NTA affinity chromatography column (Figure 6B) and quantified using the Bradford assay. A concentration of 3.6 mg/100 ml cell culture of the soluble protein was achieved under these optimal conditions.
Discussion :
RfxCas13d, also referred to as CasRx is a prominent member of the Cas13 family known for its ability to degrade lateral transcripts when targeting both abundant reporter RNAs and native RNAs. This characteristic has facilitated the use of the Cas13 family in detection of specific RNA transcripts. Techniques such as SHERLOCK have been developed utilizing this capability 12,13. Additionally, techniques like microinjection involving gRNA and the purified RfxCas13d protein into animal embryos have proven effective in silencing particular transcripts 32. The pET28b-RfxCas13d-His system (Addgene #141322) was used for expression. This system includes a kanamycin resistance gene and utilizes the T7 promoter, regulated by the lac promoter sequence, to drive RfxCas13d expression. The RfxCas13d segment of this plasmid originates from pT3TS-RfxCas13d, and the humanized RfxCas13d sequence may influence its bacterial expression" for clarity and conciseness 33. The distinct advantage of this bacterial expression plasmid over others containing RfxCas13d is its lack of a fusion partner sequence, allowing for post-expression purification through immobilized metal affinity chromatography. Previous research selected bacterial expression plasmids with the humanized RfxCas13d sequence from the Rosetta™ strain for expressing RfxCas13d 33. Rosetta™ host strains, which are derivatives of BL21, are engineered to boost the expression of eukaryotic proteins featuring infrequently used codons in E. coli. These strains carry tRNAs for AGG, AGA, CCC, CUA, AUA, and GGA codons on a chloramphenicol-resistant plasmid, facilitating a more universal translation that might be confined by E. coli codon usage 34. Nevertheless, protein expression remains an empirical process, and studies have demonstrated successful expression in BL21 strains of proteins containing rare codons 35. In this investigation, the pET28b-RfxCas13d-His plasmid was transformed into both strains, and initial observations of RfxCas13d protein expression were made. The visual findings indicated no significant differences in the target protein between the two strains. However, due to excessive non-target protein bands in the Rosetta strain, the BL21 strain was chosen for further exploration to refine expression using the RSM. In this study, we employed an RSM approach to examine how variations in IPTG concentration, temperature, OD600 nm before induction, and post-induction time impact the efficiency of RfxCas13d expression.
The ideal conditions for producing recombinant proteins are affected by multiple factors. It is important to recognize that the overexpression of proteins using plasmid DNA can impose a metabolic load, potentially disrupting growth rates and diminishing biomass accumulation. Such disturbances may lead to instability in plasmid DNA. The maximum specific growth rate is capped by the commencement of glucose overflow metabolism and the production of acetate, both of which negatively impact recombinant protein production. Consequently, it becomes crucial to establish optimal expression conditions. In this context, RSM stands out as a highly precise multivariate analysis technique capable of altering multiple parameters simultaneously 36-38. Critical factors identified for efficient recombinant protein production that require optimization, including IPTG concentration, induction timing, cell density, and temperature. Lowering IPTG concentrations can moderate transcription rates and enhance the production of soluble proteins. Typically, IPTG concentrations range from 0.1 to 1.0 mM for inducing protein expression, but reducing these concentrations further can influence solubility 39. Determining the optimal OD for induction is essential for maximizing expression levels. It is also noted that extending the incubation period post-induction does not invariably lead to increased expression levels. Over extended culture periods, proteases may degrade heterologously expressed proteins. Thus, optimizing expression duration minimizes degradation and reduces costs 40.
Traditionally, adjusting one variable at a time while holding others constant has been the approach to optimize protein expression, though this method necessitates numerous experiments and may result in misinterpreted interactions between variables. RSM offers a robust alternative to address these challenges. In the present study, the significance of the mathematical model was confirmed with a low p-value (less than 0.05). Validation tests comparing experimental outcomes with predictions from the model demonstrated a strong linear correlation, evidenced by a correlation coefficient of 0.97 and a p-value <0.05.
ANOVA results highlighted significant model terms such as Te, O, Ti, I2, Ti*Te, Te*I, O*I, O*Te, and O*Ti, with the interaction between OD600 nm before induction and time post-induction showing the most substantial impact on protein expression levels. Additionally, turbidity measurements using Image J and AlphaEaseFC software under predicted optimal conditions (IPTG concentration=250 µM, temperature= 37°C, OD600 nm before induction=0.8, and time after induction=ON) revealed a significantly high turbidity of the protein band, indicating successful expression.
In this study, the optimization of four factors- IPTG concentration, temperature, OD600 nm before induction, and time after induction-led to a significant increase in RfxCas13d expression. However; potential limitations must be acknowledged and further investigation is needed to address challenges such as the scalability of the optimized conditions and potential variations in performance when using different bacterial strains.
Conclusion :
This study establishes that all selected factors- IPTG concentration, temperature, OD600 nm before induction, and post-induction time- critically affect RfxCas13d expression. Applied RSM pinpointed the optimal values of these selection factors for producing recombinant RfxCas13d. This marks the first evaluation of the collective impact of these factors on RfxCas13d expression. Further investigation and optimization of key parameters influencing protein expression and downstream processes using RSM, along with exploring RfxCas13d expression in alternative host systems, will enhance our understanding of the ideal conditions for maximizing RfxCas13d expression.
Ethical approval :
All ethical concerns, including issues related to plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, and redundancy, were fully adhered to by the authors. The study's ethical procedures were sanctioned by the Ethics Committee of Hamadan University of Medical Sciences (Ethical code: IR.UMSHA.REC.1400.784). Written informed consent was also secured from all participants involved in the study. This research formed a part of Sepideh Abbaszadeh's Master of Science thesis at Hamadan University of Medical Sciences.
Acknowledgement :
We extend our heartfelt gratitude to the esteemed Prof. Masoud Saeedi Jam, albeit posthumously, and to our peers at Hamadan University of Medical Sciences for their contributions.
Funding: The research received financial support from the Vice-chancellor for Research and Technology at Hamadan University of Medical Sciences, Hamadan, Iran (Grant No:1400120310208).
Acknowledgement :
The authors have no relevant financial or non-financial interests to disclose.

|
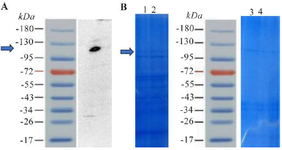
Figure 1. A) Displays the results of a Western blot analysis for the RfxCas13d protein. The recombinant RfxCas13d protein has a molecular weight of 113 kilodaltons, which aligns with the band observed in the Western blot. B) Provides a preliminary evaluation of protein expression in BL21 (DE3) and Rosetta™ (DE3) strains. Columns 1 and 2 represent expression in Rosetta™ (DE3) at 3 hr and ON post-induction, respectively, while columns 4 and 5 show expression in BL21™ (DE3) at the same time points. Protein expression levels showed no notable difference between the two strains.
|

Figure 2. Displays RfxCas13d expression in SDS-PAGE under 26 different cultivation conditions as designed by RSM. Lane M features a protein marker, while other lanes represent various experimental conditions. Turbidity values for these experiments were calculated using AlphaEaseFC software.
|
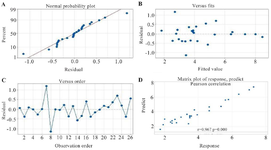
Figure 3. Diagnostic Plots for the Quadratic Models. A) Normal Probability Plot: This plot demonstrates that the residuals—the differences between observed and predicted values—are normally distributed. The alignment of points along the reference line supports the assumption of normality. B) Versus Fits Plot: This plot exhibits an ideal pattern, with residuals randomly dispersed around zero. The absence of any systematic trends or patterns indicates that the model fits the data well. C) Versus Order Plot: This plot was used to detect any time-dependent patterns or autocorrelation. The random scattering of residuals suggests no issues related to the order of observations. D) Matrix Plot of Response and Predictors: This plot presents a strong correlation matrix between the response variable and the predictors. It highlights the relationships and dependencies among the variables, indicating a well-structured model.
|
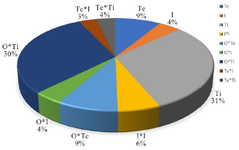
Figure 4. Evaluation of the effect of the significant parameters on RfxCas13d expression, calculated by the sum of squares index. I (IPTG concentration), Te (temperature), O (OD600 nm before induction), and Ti (post-induction time).
|
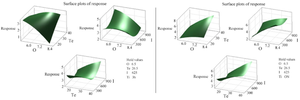
Figure 5. A three-dimensional response surface illustrates the expression of RfxCas13d. This figure examines the impact of two variables while maintaining the other two at zero levels. I (IPTG concentration), Te (temperature), O (OD600 nm before induction), and Ti (post-induction time).
|
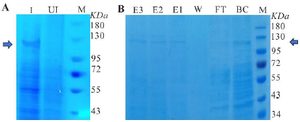
Figure 6. A) SDS-PAGE under optimal cultivation conditions is shown, induced (I), uninduced (UI), and protein molecular weight marker (M). B) RfxCas13d purification using a Ni-NTA column is shown, with eluted protein fractions (E1, E2, and E3), wash (W), flow-through (FT.), before column (BC) and protein molecular weight marker (M).
|
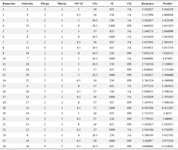
Table 1. Displays factor values and responses according to the Box-Behnken experimental design
#I (IPTG Concentration), Te (Temperature), O (Od600 nm before induction), and Ti (post-induction time).
|
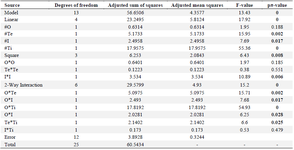
Table 2. Investigation of variance (ANOVA) for the full quadratic model concerning RfxCas13d production is detailed
# I (IPTG concentration), Te (Temperature), O (OD600 nm before induction), and Ti (post-induction time).
|
|