Recombinase Polymerase Amplification (RPA)-ELISA as an Isothermal Molecular POCT Method for Bacterial Respiratory Infection Diagnosis
-
Azizian, Reza
-
Pediatric Infectious Diseases Research Center (PIDRC), Tehran University of Medical Sciences (TUMS), Tehran, Iran
-
Biomedical Innovation and Start-up Student Association, Tehran University of Medical Sciences (TUMS), Tehran, Iran
-
Jafari, Erfaneh
-
Pediatric Infectious Diseases Research Center (PIDRC), Tehran University of Medical Sciences (TUMS), Tehran, Iran
-
Biomedical Innovation and Start-up Student Association, Tehran University of Medical Sciences (TUMS), Tehran, Iran
-
 Pourakabri, Babak
Pediatric Infectious Diseases Research Center (PIDRC), Tehran University of Medical Sciences (TUMS), Tehran, Iran, Tel: +98 21 61479; Fax: +98 21 66930024; E-mail: pourakbari@gmail.com
Pourakabri, Babak
Pediatric Infectious Diseases Research Center (PIDRC), Tehran University of Medical Sciences (TUMS), Tehran, Iran, Tel: +98 21 61479; Fax: +98 21 66930024; E-mail: pourakbari@gmail.com
-
Mamishi, Setareh
-
Pediatric Infectious Diseases Research Center (PIDRC), Tehran University of Medical Sciences (TUMS), Tehran, Iran
-
Hosseinpour Sadeghi, Reihaneh
-
Pediatric Infectious Diseases Research Center (PIDRC), Tehran University of Medical Sciences (TUMS), Tehran, Iran
-
Sotoudeh Anvari, Maryam
-
Department of Pathology, School of Medicine, Children’s Medical Center, Tehran University of Medical Sciences (TUMS), Tehran, Iran
Abstract: Background: Acute Respiratory Infections (ARIs) are a leading cause of childhood mortality worldwide, especially in African and Southeast Asian countries. Point of Care Test (POCT) techniques provide faster diagnoses compared to conventional or real-time PCR methods. Recombinase Polymerase Amplification (RPA) offers rapid on-site detection of these infections. Coupling RPA with Enzyme-Linked Immunosorbent Assay (ELISA) (RPA-ELISA) creates a cost-effective alternative, ideal for clinical applications. This study evaluates RPA-ELISA as a rapid diagnostic tool for bacterial respiratory infections.
Methods: From 11 August 2022 to 9 February 2023, respiratory samples were collected and processed using culture methods, biochemical tests, real-time PCR, and RPA assays. The RPA reactions were conducted at 39°C for 30 min, and ELISA was used for detection. Statistical analyses focused on sensitivity, specificity, Positive Predictive Values (PPV), and Negative Predictive Values (NPV).
Results: Forty-two respiratory samples, were collected in this period of which 10 samples showed no growth, and 32 tested positive. Among these positive samples, 15 isolates (35.7%) were identified as Klebsiella pneumoniae (K. pneumoniae), 14 isolates (33.3%) as Streptococcus pneumoniae (S. pneumoniae), and 3 isolates (7.1%) as Moraxella catarrhalis (M. catarrhalis). RPA-ELISA demonstrated 100% sensitivity for all pathogens, comparable to or better than RT-PCR, but had slightly lower specificity and PPV. RT-PCR achieved 100% specificity and PPV for all pathogens, indicating higher accuracy; yet, RPA-ELISA's sensitivity points to its effectiveness as a rapid screening tool.
Conclusion: RPA-ELISA is significantly faster than real-time PCR and culture methods. Its ease of use makes it suitable for on-site diagnoses in resource-limited environments. Limitations include a small sample size for certain bacteria and the necessity for further validation in varied clinical contexts.
Introduction :
Acute Respiratory Infections (ARIs) are a leading cause of mortality among children under five years of age, particularly in Low- and Middle-Income Countries (LMICs) 1,2. The World Health Organization (WHO) defines severe acute respiratory infection as an ARI accompanied by fever above 38°C, cough within the last ten days, and the need for hospitalization. ARIs encompass both upper and lower respiratory tract infections and remain a significant global health
burden, contributing to over 650,000 deaths annually in children under five 1,2. The highest incidence of ARIs occurs in African and Southeast Asian countries, where limited access to healthcare and diagnostic resources exacerbates the problem 3,4. In the United States, approximately 340,000 individuals are affected by respiratory infections each year, while in Iran, the mortality rate for infants under 27 days old due to respiratory infections is 0.4 per 1,000, rising to 4 per 1,000 for children aged 30 days to four years 5,6.
Timely and accurate diagnosis of bacterial respiratory infections is critical for effective treatment and infection control. Conventional diagnostic methods, such as culture and biochemical tests, are often time-consuming and lack sensitivity, delaying appropriate interventions 7. Molecular methods like Polymerase Chain Reaction (PCR) and real-time PCR offer improved specificity and sensitivity but require specialized equipment and expertise, limiting their use in resource-limited settings 8. The emergence of novel pathogens, such as SARS-CoV-2, and the resurgence of diseases like Ebola and Zika have further highlighted the need for rapid, portable, and cost-effective diagnostic tools 9.
Isothermal nucleic acid amplification techniques, such as Recombinase Polymerase Amplification (RPA) and Loop-Mediated Isothermal Amplification (LAMP), have gained attention for their ability to deliver rapid, sensitive, and specific results without the need for complex thermal cycling equipment 10,11. RPA, functions effectively at mild temperatures (37-42°C) and can amplify target DNA in just 20-30 min, making it an ideal choice for Point-of-Care Testing (POCT) 12. Recent research also has shown that RPA can successfully detect a variety of pathogens, such as Streptococcus pneumoniae (S. pneumoniae), Mycobacterium tuberculosis, and respiratory viruses, demonstrating sensitivity and specificity that are comparable to traditional PCR methods 13-15. Furthermore, RPA exhibits reduced susceptibility to inhibitors found in clinical samples, which simplifies the detection process and minimizes the need for extensive sample purification 16.
This study seeks to assess the efficacy of RPA combined with Enzyme-Linked Immunosorbent Assay (ELISA) (RPA-ELISA) as a rapid molecular diagnostic tool for bacterial respiratory infections. By harnessing the speed, sensitivity, and ease of RPA, this method has the potential to overcome the limitations of existing diagnostic techniques, providing a practical solution for on-site detection in resource-limited environments. The insights gained from this research could pave the way for the development of accessible and efficient diagnostic tools for ARIs, ultimately improving patient outcomes and reducing the global burden of respiratory infections.
Materials and Methods :
Individuals ranging from 1 day to 18 years old, who exhibited or were suspected to have symptoms of respiratory infections, had a fever exceeding 38°C and onset of coughing within the last ten days, were included in the study. Individuals who had taken antibiotics or antiviral medications in the past 14 days or whose predominant or primary complaint involved non-respiratory symptoms, were excluded. Bronchoalveolar Lavage (BAL), sputum, and swab samples were collected from 11 August 2022 to 9 February 2023 and divided into two parts after the preparation steps. One part was used for the gold standard of culture, and the other part was used for DNA extraction.
Due to the low frequency of Moraxella catarrhalis (M. catarrhalis) (5% and less), a duration of 20 months was considered to increase the chance of separation. After the samples were collected, they were incubated in specific environments to encourage the growth of different bacteria strains. Following this, biochemical tests were conducted to isolate the bacteria of interest. These tests included blood agar, EMB, IMViC, and M. catarrhalis specific tests. After undergoing culture and biochemical diagnosis, DNA was extracted.
Real-Time PCR: The DNA of the bacteria was separated by the columnar method, and for all three bacteria the real-time PCR method was performed. The primer and probes were designed in such a way that they could be detected at the same temperature in a reaction. Real-time PCR using the Taqman method was performed to detect the khe gene in Klebsiella pneumoniae (K. pneumoniae), the Sp2020 gene in S. pneumoniae, and the copB gene in M. catarrhalis (Tables 1 and 2) 17-21.
RPA method: When conducting RPA, it is advisable to target DNA lengths (amplicons) within the 80-400 bp range as suggested by the kit manufacturer. In a review by Lobato and O'Sullivan, DNA lengths (amplicons) were found to range from 100-200 bp 22-24. The design of RPA primers is no different from the PCR primer design, but they must be labeled with Fluorescein Amidites (FAM) in the 5-prime head and Biotin in the 3-prime head for ELISA identification (Table 2) 23-25. The RPA commercial Kit was provided by Biori Biotech Co. (Shenzhen, China) and RPA enzymes purchased from Twisdx Co. (UK). The RPA assays were carried out in a 25 µl volume using a master mix comprising 3 enzymes (Rec.A, Uvs.x, and Bst DNA polymerase) at a temperature of 39°C for 30 min. The ELISA technique was utilized for detection, taking between 45 min to an hr to complete. Overall, the RPA-ELISA method required approximately 1.15 to 1.5 hr for the entire process. In contrast, traditional culture methods typically take 24 to 48 hr to provide results, while real-time PCR methods can take anywhere from 1.5 to 2.5 hr. This indicates that the RPA-ELISA method not only shortens the diagnostic time but also minimizes the need for costly equipment, such as thermocyclers commonly used in real-time PCR.
The reaction mix contained 12.5 µl of reaction buffer, 1 µl each of Forward and Reverse primers (20 µM concentration), 1.5 µl of magnesium sulfate (20 µM), 8 µl of double-distilled water, and 1 µl of DNA (Table 3) 26.
RPA-gel electrophoresis: A 2% agarose gel was prepared for electrophoresis, and Gel Red, a safe DNA staining agent, was added to facilitate visualization. To determine the size of the amplified DNA fragments, a 100 bp DNA ladder (GeneRuler DNA Ladder Mix; SM 0331) was used as a molecular weight marker. The electrophoresis was performed at a constant voltage of 120 V for 45 min to ensure optimal separation of the DNA bands.
RPA-ELISA: For ELISA, 2 µl of the RPA product was combined with 198 µl of SCC buffer (1X; composed of 150 mM NaCl and 15 mM Sodium Citrate, pH=7), and 1.5 µl of the probe was added to this diluted solution. The diluted product was incubated at 95°C for 5 min, then transferred to the ELISA plate. Next, the plate was incubated at 37°C for 45 min and washed 3 times using the routine ELISA wash buffer. Then, 100 µl of anti-FAM-Horse Radish Peroxidase was added to the plate and incubated at room temperature for 25 min. Following this incubation, the plate was washed 3 times with the routine ELISA wash buffer. Next, 100 µl of TMB substrate was added to the plate and incubated at room temperature for 10 min. Finally, 100 µl of ELISA stop buffer was added to halt the reaction, and the results were observed 27,28.
This method can effectively identify bacterial agents qualitatively and allows for the simultaneous detection of K. pneumoniae, S. pneumoniae, and M. catarrhalis within an hr. The 3' head of the reaction product was labeled with biotin, and the 5' head with FAM. After the reaction was completed, it was poured into the ELISA wells. By connecting biotin to streptavidin at the bottom of the ELISA wells, the amplification product will be fixed at the bottom of the well. By attaching the anti-FAM antibody to the 5' end, the enzymatic reaction occurs, leading to a visible positive response upon color change of the substrate.
Statistical analysis: Statistical parameters such as True Positive (TP), False Positive (FP), True Negative (TN), False Negative (FN), Accuracy, Positive Predictive Value (PPV), Negative Predictive Value (NPV), Sensitivity, and Specificity were analyzed using SPSS version 19. Additionally, Confidence Intervals (CIs) for all metrics were calculated.
Results :
Forty-two respiratory samples, like sputum, BAL, and throat culture, were collected in the sampling period. 10 of the samples showed no growth. 15 of the identified isolates (35.70%) were K. pneumoniae, 14 (33, 34%) were S. pneumoniae, and 3 (7, 14%) were M. catarrhalis. Concentrations of RPA product for K. pneumoniae, M. catarrhalis and S. pneumoniae were 35 ng/µl, 27 ng/µl, and 32 ng/µl, respectively.
RPA-ELISA indicated better results in comparison with RPA-gel electrophoresis. The RPA-ELISA results were confirmed by real-time PCR. Real-time PCR was positive by CT: 14.2, 10.6, and 12.6 for K. pneumoniae, S. pneumoniae and M. catarrhalis, respectively. There was no band for S. pneumoniae and M. catarrhalis on gel-electrophoresis, while they indicated a positive result in RPA-ELISA reaction (Table 4). RPA-ELISA demonstrated consistent sensitivity and specificity for the specific bacteria targeted, comparable to that of real-time PCR. Sensitivity and specificity for detecting K. pneumoniae by RPA-ELISA were 100% and 80% vs. 93.3% and 100% by Real-time PCR. The PPV, NPV, and accuracy for detection of K. pneumoniae by RPA-ELISA were 40.5, 100 and 82.4%, respectively.
Sensitivity and specificity for detecting S. pneumoniae by RPA-ELISA were 100 and 80% vs. 92.7 and 100% by Real-time PCR. The PPV, NPV, and accuracy for detection of S. pneumoniae by RPA-ELISA were 38.2, 100, and 82.2%, respectively. Sensitivity and specificity for detecting M. catarrhalis by RPA-ELISA were 100 and 66.7% vs. 66.7 and 100% by Real-time PCR. The PPV, NPV, and accuracy for detection of M. catarrhalis by RPA-ELISA were 25, 100 and 70%, respectively (Table 5). Confidence Intervals for Methods for each of the bacteria are explained in (Table 6).
The RPA-ELISA method demonstrated 100% sensitivity for all pathogens tested, performing comparably or even better than RT-PCR. However, it showed slightly lower specificity and Positive Predictive Value (PPV). The total time needed for RPA-ELISA was approximately 1.15 to 1.5 hr, compared to 24 to 48 hr for traditional culture methods and up to 1.5 to 2.5 hr for real-time PCR. Nevertheless, the high sensitivity and Negative Predictive Value (NPV) of RPA-ELISA suggest it has the potential to serve as an effective rapid screening tool.
Discussion :
In this study, we developed a rapid and user-friendly detection method for K. pneumoniae using RPA. This method exhibited both 100% sensitivity and 80% specificity in clinical samples, indicating its potential as a valuable tool for the early detection of K. pneumoniae infections in hospital settings. Similarly, Tan et al developed a rapid detection platform for K. pneumoniae using RPA and CRISPR/Cas12a, achieving 100% sensitivity and specificity with clinical samples 29.
K. pneumoniae poses a significant risk to global public health, but current diagnostic methods are not conducive to rapid testing. A novel extraction-free assay named EXORCA (EXtraction-free One-pot RPA-CRISPR/Cas12a assay) has been developed for the rapid and accurate detection of K. pneumoniae based on distinct motifs 26. This test can be completed in just 30 min at a specific temperature, and the results can be observed using a fluorescence reader or with the naked eye. The feasibility of this assay was tested using 20 unextracted clinical samples, achieving a PPV of 100% (5/5) and an NPV of 100% (15/15) compared to Real-Time PCR. These results showed its potential as a point-of-care tool for pathogen identification 30.
A different study has been conducted using the autolysin gene lytA in RPA for the detection of S. pneumoniae 26. The RPA-LFS (Lateral Flow Strips) technique, employing gold nanoparticle-based lateral flow strips, demonstrated high specificity by accurately detecting 22 strains of S. pneumoniae. This method was sensitive, detecting S. pneumoniae at low levels, with results matching PCR on clinical samples. The RPA-LFS test enhanced the identification of S. pneumoniae, especially in resource-limited settings, exhibiting a 98.18% compliance rate and a kappa index of 0.977 when compared to culture-biochemistry methods 31.
Another study presented a rapid and sensitive test utilizing RPA on a disposable strip, capable of detecting S. pneumoniae and Legionella pneumophila (L. pneumophila) in less than 20 min with high sensitivity and specificity 27. The developed method is simple, suitable for near-patient and field testing, and allows for visual readout without the need for specialized equipment. In a multiplex assay amplifying specific regions of S. pneumoniae and L. pneumophila simultaneously, the analytical sensitivity was 10 Colony Forming Units (CFUs) of genomic DNA per reaction 15.
M. catarrhalis remains a neglected pathogen due to limited detection methods that hinder its diagnosis. To combat this problem, two rapid and sensitive assays, M. catarrhalis-RPA-fluorescence and M. catarrhalis-RPA-LFB (Lateral Flow Biosensor), were developed using recombinase polymerase amplification technology. These assays focus on the copB gene of M. catarrhalis and yield outcomes within 40 min, capable of detecting genomic DNA concentrations as low as 35 fg. Both assays underwent successful clinical validation using 96 Bronchoalveolar Lavage Fluid ( BALF) samples, presenting a promising solution for the quick and accurate identification of M. catarrhalis in microbiology labs and clinical settings 32.
The present study findings showed 100% sensitivity and 80% specificity for K. pneumoniae, which is consistent with other studies. Tan et al developed a rapid detection platform for K pneumoniae using RPA and CRISPR/Cas12a, achieving 100% sensitivity and specificity in clinical samples 29. Similarly, Fu et al developed an extraction-free RPA-CRISPR/Cas12a assay for K. pneumoniae, which also achieved 100% PPV and NPV, confirming this study's results 30.
For S. pneumoniae, the RPA-ELISA method demonstrated 100% sensitivity, consistent with earlier researches. Wang et al introduced a visual RPA-Lateral Flow Strip (LFS) method for S. pneumoniae, achieving high specificity and sensitivity with a compliance rate of 98.18% compared to traditional culture methods 31. Similarly, Kersting et al developed a multiplex RPA assay for S. pneumoniae and L. pneumophila, delivering high sensitivity and specificity in under 20 min, paralleling the rapid results of the current study 15.
In contrast, while our study found 100% sensitivity for M. catarrhalis, the specificity was lower at 66.7%. This aligns with challenges noted in prior studies, as M. catarrhalis has often been overlooked due to inadequate detection methods. Yu et al worked on RPA-based fluorescence and Lateral Flow Biosensor (LFB) assays for M. catarrhalis, which showed high sensitivity and specificity with detection limits as low as 35 fg of genomic DNA 32. This indicates that although the RPA-ELISA method used in the current study is effective, additional optimization may be necessary to enhance specificity for M. catarrhalis.
One of the strengths of our approach is its quick turnaround time, significantly reducing the wait for diagnostic results when compared to traditional culture methods. Additionally, the simplicity of the RPA-ELISA method makes it accessible for use in resource-limited settings. Nevertheless, a limitation of our study is the relatively small sample size for specific bacteria, such as M. catarrhalis, which may impact the generalizability of our findings. Furthermore, while the RPA-ELISA method exhibited high sensitivity, its specificity was slightly lower than that of real-time PCR. This suggests a need for further optimization to minimize false positives. Future research should also focus on determining the Limit of Detection (LOD) and Limit of Quantification (LOQ), particularly for M. catarrhalis, while increasing sample sizes and including a broader range of pathogens to enhance overall generalizability.
Conclusion :
This study presents a rapid and sensitive diagnostic method using RPA-ELISA to detect key respiratory pathogens: K. pneumoniae, S. pneumoniae, and M. catarrhalis. The assay's sensitivity and specificity are comparable to real-time PCR, offering faster results, ease of use, and minimal equipment needs, making it suitable for point-of-care and resource-limited environments. It achieved 100% sensitivity and negative predictive value for all pathogens, demonstrating reliability in ruling out infections. However, specificity for M. catarrhalis was lower, influenced by small sample sizes and low pathogen prevalence.
Despite these challenges, the assay has great potential for early detection and treatment of respiratory infections, addressing significant public health concerns. Further validation in diverse clinical settings is recommended to assess its broader applicability.
Acknowledgement :
We would like to express our sincere gratitude to Dr. Hesam Kopahi for their invaluable guidance and feedback throughout this study. We are also grateful to Dr. Sadegh Zomorodimanesh for their valuable input and assistance. And our appreciation also goes to Diazist and Pishgaman Sanjesh Isatis for their collaboration and contributions. The sampling protocol was approved by the Ethical Committee of Tehran University of Medical Sciences, Tehran, Iran (IR.TUMS.CHMC. REC.1401.057).
Conflict of Interest :
Authors declare no conflict of interest.
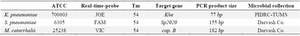
Table 1. Data of Real-time
Pediatric Infectious Disease Research Center (PIDRC), Tehran University of Medical Sciences (TUMS).
|
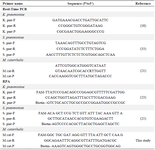
Table 2. The primers and probes used in this study
- pne: K. pneumoniae; S. pne: S. pneumoniae; M.cat: M. catarrhalis.
|
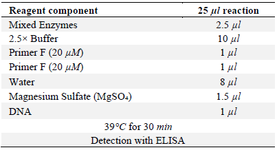
Table 3. RPA protocol reaction
|
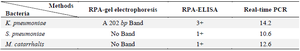
Table 4. Comparison between RPA-ELISA, RPA-gel electrophoresis and real-time PCR
|
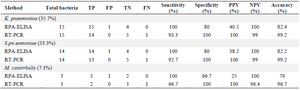
Table 5. Comparison between RPA-ELISA’s sensitivity and specificity with real-time PCR
Each bacteria’s prevalence has been shown in parenthesis; TP: True Positive; FP: False Positive; TN: True Negative; FN: False Negative; PPV: Positive Predictive Value; NPV: Negative Predictive Value.
|
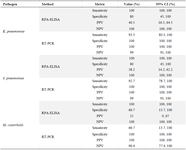
Table 6. Comparison between RPA-ELISA’s metric based on CI
|
|