Study of DACH1 Expression and its Epigenetic Regulators as Possible Breast Cancer-Related Biomarkers
-
Nasirpour, Mohammad Hossein
-
Department of Medical Genetics, Institute of Medical Biotechnology, National Institute of Genetic Engineering and Biotechnology (NIGEB), , Tehran, Iran
-
 Salimi, Mahdieh
Department of Medical Genetics, Institute of Medical Biotechnology, National Institute of Genetic Engineering and Biotechnology (NIGEB), Tehran, Iran, Tel: +98 21 44787382; Fax: +9821 44787395; E-mail: salimi@nigeb.ac.ir
Salimi, Mahdieh
Department of Medical Genetics, Institute of Medical Biotechnology, National Institute of Genetic Engineering and Biotechnology (NIGEB), Tehran, Iran, Tel: +98 21 44787382; Fax: +9821 44787395; E-mail: salimi@nigeb.ac.ir
-
Majidi, Faezeh
-
Department of Biology, Science and Research Branch, Islamic Azad University, Tehran, Iran
-
Minuchehr, Zarrin
-
Institute of Industrial and Environmental Biotechnology (IIEB), National Institute of Genetic Engineering and Biotechnology (NIGEB), Tehran, Iran
-
Mozdarani, Hossein
-
Department of Medical Genetics, Faculty of Medical Sciences, Tarbiat Modares University, Tehran, Iran
Abstract: Background: Breast carcinogenesis involves both genetic and epigenetic changes. DNA methylation, as well as micro-RNA regulations, are the significant epigenetic phenomena dysregulated in breast cancer. Herein, the expression of DACH1 as a tumor suppressor gene and its promoter methylation status was analyzed in breast cancer tumors. Also, the expression of three micro RNAs (miR-217, miR-6807-3p, and miR-552), which had been previously reported to target DACH1, was assessed.
Methods: The SYBR green-based Real-Time reverse transcription-PCR was used to determine DACH1 and micro-RNAs (miR-217, miR-6807-3p, and miR-552) expression in 120 ductal breast cancer tumors compared with standard control. Also, the promoter methylation pattern of DACH1 was investigated using the Methylation-specific PCR technique.
Results: DACH1 expression was significantly down-regulated in breast tumors (p< 0.05). About 33.5% of tumors showed DACH1 promoter hyper-methylation. The studied micro-RNAs, expression was negatively correlated with DACH1 expression. The highest expressions of miRNAs and higher DACH1 promoter methylation were observed in advanced cancer situations. The Kaplan-Meier survival curves indicated that the overall survival was significantly poor in higher miRNAs and lower DACH1 expression in breast cancer patients (p<0.002).
Conclusion: DACH1 down-regulation may be associated with a poor breast cancer prognosis. The DACH1 down-regulation may be due to epigenetic regulations such as promoter methylation, especially in triple-negative cases. Other factors, such as micro-RNAs (miR-217, miR-6807-3p, and miR-552), may also have an impact. The elevated expression of miR-217, miR-6807-3p, and miR-552, maybe candidates as possible poor prognostic biomarkers in breast cancer management for further consideration.
Introduction :
Breast cancer is the second most common cause of cancer-related death and the most frequent cancer in women worldwide 1. Despite advances in screening programs and better education that have led to a significant decline in breast cancer mortality, unfortunately, about 25-40% of breast cancers lead to metastases and death 1. Unfavorable clinical results are primarily due to metastasis, therapy resistance, and tumor recurrence. Identifying new biomarkers related to aggressive phe-notypes, molecular subtypes, and prognosis of breast cancer is essential for drug development, proper treatment, disease surveillance, and cancer management. Therefore, understanding the molecular bases of such a diverse behavior of breast cancer subtypes and investigating novel and more effective therapeutic strategies is urgently needed 2.
Epigenetic mechanisms such as DNA methylation, histone modifications, long non-coding RNA, and micro-RNA regulations have emerged as fundamental factors in breast cancer development and progression 3. The epigenetic modifications are believed to be early events in breast cancer development due to their presence even in carcinoma in situ lesions, which makes them very suitable as early detection biomarkers 4.
Dachshund family transcription factor 1, or Dachshund homolog 1 (DACH1), as a critical cell fate determination factor, plays a vital role in breast cancer 5. DACH1 is expressed in normal human breast epithelium, while down-regulation of DACH1 protein was found in many human breast cancers, significantly correlated with poor prognosis 6. However, the reintroduction of DACH1 indicatively reversed the transformed phenotype in these human mammary epithelial cells with some oncogenes 6,7. In previous studies, DACH1 was reported to be epigenetically silenced in colorectal cancer and clear cell renal cell carcinoma 8,9. However, the mechanism underlying the frequent loss of DACH1 in breast cancer has not been well explored yet.
In breast cancer, promoter hypermethylation has been reported in more than a hundred genes that most of them play essential roles in cell-cycle regulation, DNA repair, regulation of cell transcription, apoptosis, cell adhesion, hormone-mediated cell signaling, and tissue invasion and metastasis 10,11. In addition to hypermethylation, hypomethylation also represents a common feature of breast tumors and frequently occurs in regions of segmental duplications 11.
Several lines of evidence have demonstrated that the down-regulation of DACH1 is closely associated with the proliferation, invasion, and metastasis of various tumors, including breast 7,12, lung 13, esophageal cancer 14, renal cell 15, and hepatocellular carcinoma 16. Micro-RNAs (miRNAs) are another main epigenetic element that has an essential role in gene regulation. The miRNAs are short (18-25 nucleotides), highly conserved non-coding RNAs, which perform a significant function in biological stability and gene expression at a post-transcriptional level by translation inhibition or degradation of protein-coding mRNAs via imperfect base-pairing to complementary sites on target mRNAs 17.
The novel aspect of cancer studies comes from the fact that the micro RNAs can adjust the levels of their corresponding mRNAs and serve critical roles in the pathological and physiological processes of tumor development. The possibility that miRNA may be a novel focus for examining the current diagnosis and treatment of tumors has taken power 17. The miR-552, miR-217, and miR-6807-3p are three miRNAs that have already been reported to directly target DACH1 mRNA 18-20. About miR-217 and its association with cancer, conflicting results have been reported. Some studies have reported that down-regulation of miR-217 was strongly associated with tumor migration and invasion in malignancies such as hepatocellular carcinoma 21 and gastric cancer 22. In contrast, it was reported that miR-217 promotes tumor proliferation in breast cancer by targeting DACH1 19. Also, it inhibits triple-negative breast cancer cell growth, migration, and invasion by targeting KLF5 23.
MiR-552 is a newly discovered miRNA; its function and mechanism of action in biological processes and diseases are not entirely understood 24. Previous studies showed that miR-552 promotes colorectal cancer cell proliferation and migration by directly targeting DACH1 via the Wnt/β-catenin signaling pathway 18,25. The up-regulation of miR-552 was reported in ovarian cancer, especially in metastasis and recurrence 24. Also, miR-552 regulates liver tumor-initiating cell expansion and Sorafenib resistance 26. However, the role of miR-552 in breast cancer was little known.
Another micro-RNA studied in the present research is miR-6807-3p, which has been reported as a novel miRNA that promotes tumor-promoting influences on glioma progression by directly targeting DACH1 20. Here the DACH1 expression and its possible epigenetics regulators focusing on its promoter methylations and expression of a panel of micro RNAs targeting DACH1 (miR-217, miR-552, and miR-6807-3p) were studied in breast cancer patients.
Materials and Methods :
Study population: This study was a case-control study. 120 tumor and standard adjacent tissue samples were obtained from breast ductal carcinoma females, and 40 samples of women with no cancer history underwent surgery due to mammoplasty. The samples were collected from the patients admitted to Imam-Khomeini Hospital (Tehran, Iran) between 2016 and 2017. The protocol was based on the Helsinki declaration, and patients and controls signed a written informed consent letter before enrollment (approval registered 52D/4922 dated 29 September 2016). Diagnosis of ductal carcinoma and availability of pathologic diagnostic information were considered inclusion, and receiving any chemo/radiotherapy before recruitment and any history of familial malignancy were considered exclusion criteria. The demographic and histochemical data of the patients and controls are summarized in table 1. Tumor staging was performed according to the Tumor, Node, and Metastasis (TNM) classification based on the American Joint Committee on Cancer (AJCC) staging system.
RNA and DNA extraction: Tissue samples (both tumor and normal) were immediately snap-frozen and stored at -70°C until use. Total RNA was extracted and purified from fresh-frozen biopsies with the RNX- Plus Kit (CinnaGen Co., Iran, EX6101), a guanidine/ phenol solution, for total RNA isolation from the homogenized sample. Through the action of guanidine salt in the RNA isolation procedure, DNA and protein were precipitated in the phenol phase, and the aqueous phase contains high-quality all types of genomic RNA. DNA has been extracted by the manufacturer’s protocol using a DNeasy Blood and Tissue kit (Qiagen, Hiden, Germany, 69504). The RNA and DNA concentration and purity were confirmed using absorbance measurements with a NanoDrop 2000 (Thermo Scientific, USA).
Real-time reverse transcriptase (RT) -PCR: Real-time RT-PCR was performed to detect the expression levels of the DACH1 gene and micro-RNAs (miR-217, miR-6807-3p, and miR-552) after cDNA synthesis using Precision qScriptT Reverse Transcription Kit (Primerdesign, Chandlers’s Ford, UK, RT-NanoScript2) for mRNA expression and BonmiR cDNA kits (Bon biotech, Iran, BN-0011.17.2) for micro RNAs. All the steps were done according to the manufacturer’s instructions. Beta-actin and SNORD47 were used as the internal controls for DACH1 and micro-RNAs, respectively. The primer sequences for DACH1 and beta-actin were as follows: DACH1: F: 5′ ATGTGGAACAAGTTCGCATCC 3′ and R: 5′ TGCA GTCATTGTAGAGGGTCT 3′; Beta-actin: F: 5′ GAGACCTTCAACACCCCAGC3′ and R: 5′ AGACGCA GGATGGCATGG 3′. All PCRs were performed using a Light Cycler TM system (Rotor gene, Corbett, Germany). For each sample, 500 ng/µl of total cDNA was used. cDNA was mixed with 0.3 µM of each forward and reverse primer with 10 µl of SYBR green master mix (Roche, Germany, 4707516001) to a final reaction volume of 20 µl. The thermal cycling conditions comprised an initial denaturation step at 95°C for 7 min and 45 cycles at 95°C for 10 s and 60°C for 30 s, and 72°C for 20 s. Experiments were performed with triplicates for each data point.
The primer set for micro-RNA expression analysis for designing and synthesis was ordered from Stem Cell Technology Company (Tehran, Iran). The ordered set comprised miR-217, miR-6807-3p, miR-552 specific forward primer, universal reverse primer, and internal control forward primer (SNORD47). The thermal cycling conditions consist of an initial denaturation step at 95°C for 2 min and 40 cycles at 95°C for 5 s and 60°C for 30 s. Amplification efficiency for each primer pair was determined by amplifying a standard linear curve (from 0.10 to 1,000 ng) of total cDNA assessed by an ultraviolet spectrophotometer. The standard curves have shown good linearity and amplification (approximately 100%). The expression level was calculated by the 2-△△CT method. The data were presented as the fold change in gene expression normalized to an endogenous reference gene relative to the controls. The two-fold and more RNA expression is considered up-regulation, between 0.5- and 2-fold as usual and 0.5-fold and less as down-regulation.
Methylation-specific PCR (MS-PCR): Bisulfite treatment of isolated DNA was performed using the EpiTect Bisulfite Kit (Qiagen, Hilden, Germany, 59104) according to the manufacturer’s instructions. The methylation status of the DACH1 gene was determined qualitatively by the Methylation-Specific Polymerase Chain Reaction (MS-PCR). Primer sequences used for MS-PCR analysis, with PCR product size and primer annealing temperature, are listed as follow: DACH1 methylated-specific forward primer was 5'-GGAAAAAATTATTAGTTTTC GCGGAC-3' with the annealing temperature of 61.6°C, and reverse primer, 5'- AAACCGAAAA CACAAAAATAACGA TC-3' with annealing temperature of 60°C, and DACH1 un-methylated forward primer: 5'- TTTGGAA AAAATTATTAGTT TTTGTGGAT-3' with annealing temperature of 59.1°C, and reverse 5'- AAAAAACCAAAAACACAAAAATAACAATCA-3' with annealing temperature of 59.8°C, the amplicon size was 130 base pair length. Four µl of bisulfite-modified DNA were subjected to PCR amplification in a final reaction volume of 25 µl comprised of 12.5 µl of 2× EpiTect MSP Kit master mix (Qiagen, Hilden, Germany, 59305) and 0.5 µM of each primer. PCR was performed with an initial 10 min of incubation at 95°C, followed by 40 cycles of denaturation, 30 s at 95°C, 30 s of annealing at 60°C, extension at 72°C for 60 s, and a final 10 min of hold at 72°C as a final extension phase. Each sample was assessed in duplicate, and each run included no template control (NTC) and the EpiTek PCR control DNA set (methylated and un-methylated DNA) as external universal control (Qiagen, Hilden, Germany, 59655, 59665). Aliquots of PCR products were separated on 1.5% agarose gel, stained with ethidium bromide, and appeared under UV illumination.
Statistical analysis: Data were analyzed using SPSS 16.0 (SPSS Inc. Chicago, USA). The Mann-Whitney U test/Kruskal-Wallis test and the Chi-square test were performed for numerical and parametric data, respectively. Correlation and consistency were analyzed using Pearson correlation analysis. The Receiver Operating Characteristic (ROC) curve was used to illustrate diagnostic ability. Kaplan–Meier survival analysis was also performed. Numerical data are presented as the mean ± Standard Deviation (SD). Differences were considered statistically significant if p<0.05.
Results :
mRNA expression levels of the DACH1 gene: The mean relative expression of DACH1 normalized with the beta-actin gene in breast tumor tissues was 0.17±0.27, indicating this gene’s down-regulation com-pared with normal control groups. As shown in figure 1, the DACH1 mRNA expression in breast cancer patients showed a wide range from 0.001 to 0.89 fold changes. The data revealed that about 12.5% of the patients showed normal DACH1 expression, whereas most patients (about 82.5%) were DACH1 gene down-regulated in breast tumors. As indicated in table 2, there was no statistically significant association between DACH1 gene expression and Human Epider-mal Growth factor Receptor 2 (HER2) or hormone receptor status (estrogen receptor, progesterone receptor) (p> 0.05). However, in the Triple-Negative (TN) breast tumors, the DACH1 expression was significantly lower than the non-triple negative ones (p=0.0003) as well as a significant decrease in DACH1 expression was observed in metastatic and LN + (lymph node involved) breast cancer cases (p<0.0001). The data also indicated the significant down-regulation of the DACH1 gene in the advanced stages of the disease (third and fourth stages) (p=0.0012).
As shown in figure 2, the ROC curve analysis was performed between the breast cancer patients focusing on both the lymph nodal status (Figure 2A) and being (triple negative) TN or non-TN (Figure 2B). The ROC curve of DACH1 expression for discriminating LN+ breast cancer patients from LN negatives had an area under the curve of 0.93 (95% confidence interval= 0.87-1) (Figure 2A), and for discriminating TN from non-TN breast cancer patients had the area under the curve of 0.89 (95% confidence interval=0.80-0.98), (Figure 2B).
All patients were followed up for at least five years. The Overall Survival (OS) curves in lower and higher than mean DACH1 expression groups were shown in figure 3A, and patients with lower DACH1 expression displayed a poor OS (p=0.0017).
DACH1 gene promoter methylation status: The methylation status of breast tissues comprised of the tumor, normal adjacent, and normal control are summarized in table 3. The data indicated that 30% of breast tumors showed methylation of the DACH1 promoter region, significantly higher than normal and normal adjacent control. Considering the frequency of the un-methylated promoter of the DACH1 gene, which was 68.3%, it could be concluded that promoter methylation may not be the only factor that has an impact on the expression regulation of the DACH1 gene.
About 86.7% of the normal tissues adjacent to the tumor were un-methylated in promoter regions of the DACH1 gene. In comparison, 13.3% of the normal tissues adjacent to the tumor simultaneously showed a dual pattern, both methylated and non-methylated. The frequency of DACH1 promoter methylation was assessed in different breast cancer patient groups based on clinicopathological characteristics, as shown in table 4. The data revealed that the highest frequency of DACH1 promoter methylation was observed in poor prognostic cancer indicator groups, triple-negative and stage 4 with distant metastasis. There were no statistically significant differences between other clinicopathological characteristics such as being estrogen receptor positive or negative (ER+/ER-), progesterone receptor positive or negative (PR+/PR-), human epidermal growth factor receptor two positive or negative (HER2+, HER2-) or lymph-node involvement or not (LN+/LN-) (p>0.05).
Expression of miR‑217, miR-6807-3p, and miR‑552 in the test and control groups: As shown in figure 4A, the expression levels of all three miRNAs (miR‑217, miR-6807-3p, and miR‑552) were significantly up-regulated in breast cancer patients compared with the control group (p<0.0001). There were no statically significant differences between normal adjacent and normal control groups in miRNAs expression (data are not shown). The expression of studied miRNAs was evaluated in different clinicopathological features of breast cancer patients in the study group. The experimental results revealed that miR‑217, miR-6807-3p, and miR‑552 expression in the test group was related to higher stages of the disease and lymph nodal involvement, which are both considered poor prognosis indicators (p<0.0001, Figures 4B and C). The other factors, such as hormone and HER2 receptor status, were not positively associated with miRNA expression. The ROC curve analysis between studied miRNAs expression and lymph nodal involvement, focusing on the area under the curves, indicated the importance of miRNAs expression in metastasis (Figure 5). The negative correlation between the expression of DACH1 and all three miRNAs (miR‑217, miR-6807-3p, and miR‑552), r= -0.99, stated that the expression of the studied miRNAs (miR‑217, miR-6807-3p, and miR‑552) and DACH1 gene is inversely related. All patients were followed up for at least five years. The OS curves in lower and higher than the mean expression of all three studied miRNA (miR‑217, miR-6807-3p, and miR‑552) groups were shown in figure 3B, and patients with higher simultaneous expression of three miRNAs displayed a poor OS (p= 0.0001).
Discussion :
The epigenetic dysregulation associated with genetic alterations is a component of breast cancer. The genomic instability due to epigenetic pathogenesis is central to breast cancer hallmarks. Being plastic and reversible, epigenetic processes appear more amenable to therapeutic intervention than the more unidirectional genetic aberrations 27. DACH1 plays a vital role in the natural evolution of several organs as a member of the Retinal Determination Gene Network (RDGN), consisting of other genes such as EYA1 and SIX1. The DACH1 gene acts as a tumor suppressor, and its reduced expression can lead to cancer 28. Many observations have shown that down-regulation of the DACH1 gene leads to the proliferation of cancer cells, invasion, and metastasis in a variety of malignancies, including mammary 7,12,29, lung 13, esophageal 14, renal 15, and liver 16 tumors. There is strong evidence that nuclear DACH1 expression can act as a tumor suppressor gene. Nuclear DACH1 protein expression was significantly associated with markers of good prognosis, including low cell proliferation (MIB1 expression) and functional apoptosis (Bcl2 expression). It has previously been observed that reduced DACH1 expression occurs in invasive breast cancer 30. This finding was confirmed by our data, where DACH1 face showed an inverse association with poor prognosis indicators such as nodal and distant metastasis and higher cancer stages.
More recently, it was reported that DACH1 inhibited the invasion and metastasis of breast cancer cells by decreasing MMP9 expression. Regarding mechanism, DACH1 interacted with p65 and c-Jun at the NF-κB and AP-1 binding sites, respectively. Moreover, DACH1 reduced the acetylation level of p65 by recruiting HDAC1 and then repressed the transcriptional activity of p65 7. Further evidence of its tumor suppressor gene function is that DACH1 homozygous deletion stimulates tumorigenesis in glioma cells 31, and loss of DACH1 occurs in the high FIGO surgical stage endometrial cancers 32. DACH1 represses steroid hormone signaling by interacting with hormone receptors, including ER and androgen 33,34. Cytoplasmic DACH1 expression levels also correlated to several clinicopathological parameters, including histologic grade, FIGO surgical score, and pathological type. Again, cytoplasmic expression of DACH1 was more prevalent in higher-grade tumors like G2-3/G3 versus G1 to G2 and stage III to IV versus stage I to II 35. Moreover, the nuclear expression of DACH1 in ovarian cancer correlated with histologic grade and FIGO surgical stage. Finally, a comparison between the expression of DACH1 and one of the major players in ovarian cancer, ER+, revealed that DACH1 localization was preferentially cytoplasmic in the ER-positive ovarian cancer samples 35. Additionally, DACH1 has been identified to inhibit the transforming growth factor‑β CRC signaling pathway in ovarian and breast cancer 36 and to restrain the Wnt/β‑catenin signaling pathway in Colorectal Cancer (CRC) 15. Also, it affects the PI3K/AKT signaling pathway, a vital kinase cascade that plays a role in the regulation of cellular quiescence, proliferation, cancer, and longevity 37, and its disruption can lead to tumorigenesis.
The Wnt/β‑catenin pathway, as an evolutionarily conserved pathway, is essential in initiating and regulating a diverse range of cellular activities, including calcium homeostasis, cell proliferation, and cell polarity 38. Several target proteins are in the Wnt/β‑catenin signaling pathway, including cyclin D, c‑Myc, MMP3, and LEF. Previous studies have revealed that DACH1 exhibited an inverse correlation with cyclin D1 and c‑Myc 8. It was reported that DACH1 might exert inhibitory effects on the development of breast cancer partly by suppression of EMT inducers and CSCs markers, especially CD44 5.
This study showed that the expression of the DACH1 gene in tumor tissue was significantly reduced compared to its typical adjacent normal breast tissues and normal control. In fact, in 82.5% of tumor tissues, there was a reduction in DACH1 gene expression. These data confirm the possible role of the DACH1 gene as a tumor suppressor. Although tumors are caused by the accumulation of a series of genetic changes, the formation of tumors is not only limited to genetic differences, epigenetic changes could also contribute to this phenomenon. Epigenetics is a mechanism for the stable maintenance of gene expression that involves physically 'marking' DNA or its associated proteins, which allows genotypically identical cells to be phenotypically different 39. Epigenetic marking of the genome has several forms. Acetylation, phosphorylation, ubiquitylation, and methylation of histones and DNA methylation are potential mechanisms for the epigenetic tagging of the genome 40,41. DNA methylation, as the most prominent epigenetic marker, refers to adding a methyl (CH3) group on the cytosine arm in the genome-rich CpG nucleotide regions known as CpG islands 42. Studies have shown that hypermethylation in the CpG islands in the promoter regions of tumor suppressor genes prevents them from being expressed and thus can promote carcinogenesis 27.
In the present study, the data revealed that about 32% of the breast tumor samples had methylation in the promoter region of the DACH1 gene. However, in the normal adjacent tissues (average margin of the tumor), about 87% of the cases lacked methylation in the promoter region of this gene, and about 13% of the cases simultaneously yielded both bands corresponding to methylated and non-methylated promoters. Methylation of the DACH1 gene promoter was not observed in normal breast tissue extracted from women who underwent cosmetic surgery.
Based on histopathological characteristics, the study of DACH1 promoter methylation frequency in different groups of breast cancer patients indicated a positive association between DACH1 promoter methylation and distant metastasis and triple negative hormone receptor status (p<0.001). In this study, the association between the higher stages of breast cancer and the more significant methylation of the DACH1 gene was observed, which suggests further suppression of this tumor suppressor in the advanced stages of cancer.
It could be concluded that DACH1 as a tumor suppressor gene may have a role in breast cancer induction, but the reduction of DACH1 gene expression does not seem to be solely due to the promoter's methylation. According to several previous reports, interactions between miRNAs and their corresponding mRNA regulate the expression of proto-oncogenes or tumor suppressor genes 43 and determine the malignant characteristics of tumor cells, and were involved in almost every step during the initiation, development, and progression of breast cancer 44.
Numerous studies have reported miRNA dysregulation in cancerous cell growth and proliferation, cell death inhibition, immune invasion, neoangiogenesis, and metastasis 45. Also, miRNAs regulate the fate of cancer stem cells (CSCs) 27. The cancer miRNA dy-sregulation may result from the alteration of epigenetic and genetic regulation of the miRNA genes, aberrant miRNA biogenesis, and disruption of miRNA transcriptional control 45. The copy number variation due to the amplification or deletion of miRNA genes, arising from the cancer-associated genomic instability, the hy-permethylation of CpG islands near the miRNA genes, and epigenetic drugs have been reported to alter the expression profile of miRNAs 46. It was reported that miRNA dysregulation is associated with the progression through every stage of breast cancer to metastasis 27. Also, miRNAs provide notable information for breast cancer diagnosis, subtyping, prognosis, and treatment monitoring 27.
In the present study, the expression of miR-217, miR-6807-3p, and miR-552, the three miRNAs which had been already reported to target DACH1 directly, were studied 18-20. The present data revealed that the expression of the three miRNAs (miR-217, miR-6807-3p, and miR-552) was significantly upregulated in breast cancer tumors compared with normal control. Notably, these data indicate that high expression of these miRNAs was associated with the proliferation and migration of breast cancer. The present study may explain the role of miR-217, miR-6807-3p, and miR-552 in breast cancer.
Previous studies confirmed our findings in several cancers. For instance, increased expression of miR-552 acts as a potential predictor biomarker for poor prognosis of colorectal cancer by directly targeting DACH1. In addition, miR-552 also enhances the metastatic capacity of colorectal cancer cells by targeting metalloprotease and disintegrin 28. It was also reported that miR-552 over-expression correlated well with the poor prognosis of ovarian cancer patients 24.
miR-217 was reported as a novel onco-miR in breast cancer. It could accelerate the cell cycle progression by directly targeting the DACH1 gene and inhibiting its anti-cancer functions and proposed inhibiting miR-217 as a potent therapeutic strategy for breast cancer 19. In contrast, in renal cell carcinoma, the lower expression of miR-217 was reported to be associated with higher tumor grade and stage and stated that the down-regulated expression of miR-217 was associated with tumor development and metastasis 22.
miR-6807-3p was reported as a new miRNA that promoted cell proliferation and migration, inhibited apoptosis in glioma cells, and served as an oncogene in a miR-217-5p- independent manner in glioma. It was reported that miR-6807-3p could be an essential tumor facilitator in vitro. DACH1 was registered as a target molecule of miR-6807-3p 20.
Conclusion :
The data indicate that reducing DACH1 gene expression may be associated with a poor breast cancer prognosis. This reduction may be due to epigenetic regulations such as promoter hypermethylation, especially in triple-negative cases, and micro-RNAs (miR-217, miR-6807-3p, and miR-552) up-regulation. The data revealed that higher expression of the studied micro-RNAs (miR-217, miR-6807-3p, and miR-552) may be considered poor prognosis possible biomarkers in breast cancer management.
Acknowledgement :
We sincerely thank all the individuals who participated in this study. This project was supported by the National Institute of Genetic Engineering and Biotechnology (NIGEB) (Grant number is 961202-I-651, ethical registration code is 52D/4922 dated 29 September 2016).
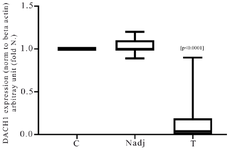
Figure 1. The Real-time RT-PCR analysis of DACH1 mRNA expression in breast cancer tumors (T) and normal adjacent margin of the tumors (Nadj) compared with normal control (C). Results are expressed as fold number changed versus control. The DACH1 RNA values were previously normalized to beta-actin.
|
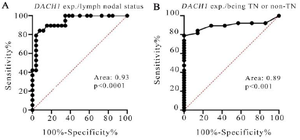
Figure 2. The ROC curve of DACH1 expression between the breast cancer patients discriminating both the lymph nodal status (Figure 2 a) and being TN or non-TN (Figure 2 b).
|
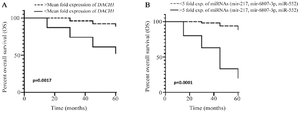
Figure 3. Kaplan–Meier survival curves of breast cancer patients based on DACH1 expression status (p=0.0017) (A) Kaplan–Meier survival curves of breast cancer patients based on the simultaneous expression of three studied miRNA (miR‑217, miR-6807-3p, and miR‑552) status (p=0.0001) (B).
|

Figure 4. The Real-time RT-PCR analysis of three miRNAs (miR‑217, miR-6807-3p, and miR‑552) expression in breast cancer tumors compared with normal control (a), between different stages of the disease (b) and between LN-, and LN+ tumors (c). Results are expressed as fold number changed versus control. The miRNA values were previously normalized to SNORD47.
|

Figure 5. The ROC curve of miRNAs expression between the breast cancer patients discriminating the lymph nodal status, A) miR-552, B) miR-6807-3p, C) miR-217.
|

Table 1. Characteristics of breast cancer patients and controls
ER: the Estrogen Receptor, PR: Progesterone Receptor, HER2: Human Epidermal Growth Factor Receptor, LN: Lymph node, IHC: Immunohistochemistry, CISH: Chromogenic in-situ hybridization, FISH: Fluorescence in situ hybridization.
|
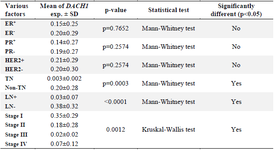
Table 2. The mRNA DACH1 expression in breast cancer tumors is stratified according to the clinicopathological character
ER: the Estrogen Receptor, PR: Progesterone Receptor, HER2: Human Epidermal Growth Factor Receptor, LN: Lymph Node, TN: Triple-Negative (ER-, PR-, HER2-).
|

Table 3. Categorization of DACH1 promoter methylation status
|
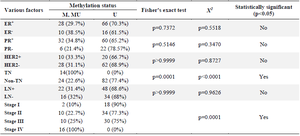
Table 4. DACH1 promoter methylation status in different breast cancer groups based on hormone receptors status, lymph node involvement, and various stages of breast cancer
M: Methylated DACH1 promoter, U: Un-methylated DACH1 promoter, MU: Both methylated and Un-methylated DACH1 promoters.
|
|