Association between PTCH1 and RAD54B Single-Nucleotide Polymorphisms and Non-syndromic Orofacial Clefts in the Northeast Population of Iran
-
Morvaridi Farimani , Reza
-
Department of Orthodontics, School of Dentistry, Shahid Beheshti University of Medical Sciences, Tehran, Iran
-
Azimi-Nezhad , Mohsen
-
Non-Communicable Diseases Research Center, Neyshabur University of Medical Sciences, Neyshabur, Iran
-
UMR, INSERM U 1122, IGE-PCV, Interaction Géne-Environment Enpathophysiologie Cardiovasculaire, Université De Lorraine, Nancy, France
-
 Ebadifar , Asghar
Department of Orthodontics, School of Dentistry, Shahid Beheshti University of Medical Sciences, Tehran, Iran, Tel: +98 21 26708362; Fax: +98 21 22403194; E-mail: a.ebadifar@sbmu.ac.ir
Ebadifar , Asghar
Department of Orthodontics, School of Dentistry, Shahid Beheshti University of Medical Sciences, Tehran, Iran, Tel: +98 21 26708362; Fax: +98 21 22403194; E-mail: a.ebadifar@sbmu.ac.ir
-
Dentofacial Deformities Research Center Research Institute of Dental Sciences, Faculty of Dentistry, Shahid Beheshti University of Medical Sciences, Tehran, Iran
-
Jafarian , Zahra
-
Iranian Research Center on Aging, University of Social Welfare and Rehabilitation Sciences, Tehran, Iran
-
Kamali, Koorosh
-
Department of Public Health, Faculty of Public Health, Zanjan University of Medical Sciences, Zanjan, Iran
-
Nazari , Zeinab
-
Non-Communicable Diseases Research Center, Neyshabur University of Medical Sciences, Neyshabur, Iran
Abstract: Background: Non-Syndromic Cleft Lip with or without cleft Palate (NSCL/P) is a common developmental disorder of the head and neck with a multifactorial etiology. The current study aimed to evaluate the potential association of PTCH1 (rs10512248) and RAD54B (rs12681366) polymorphisms with NSCL/P in the Northeast Iranian population.
Methods: In the present study, blood samples were taken from 122 subjects with NSCL/P and 161 healthy controls. Polymerase Chain Reaction (PCR) followed by Restriction Fragment Length Polymorphism (RFLP) were used to conduct genotyping of single-nucleotide polymorphisms.
Results: Although differences were observed between cases and controls in rs10512248 and rs12681366, our data did not support a significant association of these polymorphisms with NSCL/P in our population.
Conclusion: Our findings suggest that polymorphisms of rs10512248 and rs12681366 may not be potential risk factors for NSCL/P in the Northeast Iranian population due to the multifactorial and multiethnicity characteristics of some genes.
Introduction :
Disregarding the different forms of heart deformities, orofacial clefts are the most frequent congenital anomalies in humans 1,2. Whether the patients have other anomalies or malformations, these clefts can be classified into syndromic and non-syndromic forms 3. However, the incidence and presentation of these congenital disorders vary broadly by ethnic origin, geographical position, and socioeconomic status 4,5. According to previous reports, the prevalence of Non-Syndromic Cleft Lip with or without cleft Palate (NSCL/P) was much higher in some ethnic groups, including Asians and Native Americans, followed by Caucasians and Africans 6. Although there is no precise evaluation of the NSCL/P population in Iranians, the approximate incidence of NSCL/P is about 1 per 1000 live births 7.
NSCL/P has a multifactorial etiology that both genetic and environmental factors contribute to disease susceptibility 8. Over the recent years, using genetic association studies, several susceptible genes have been identified to have a role in the complex etiology of NSCL/P 9. In recent years, Genome-Wide Association Studies (GWAS) and Linkage Disequilibrium analysis have identified several pathogenic genes such as MTHFR, FGF1, MSX1, IRF6, VAX1, and SUMO1 to associate with NSCL/P 10. Recently, GWAS has confirmed that the SNPs of PTCH1 (rs10512248) and RAD54B (rs12681366) are associated with NSCL/P in different populations, including Chinese, Africans, and Irish 11-13.
PTCH1 is located on chromosome 9q22.3 and is responsible for producing the patched-1 protein, a receptor for the sonic hedgehog ligand 14. Patched-1 and Sonic Hedgehog are part of a complex process that defines the form of different parts of the developing human body and craniofacial morphogenesis 10. Previous research on mice has displayed that PTCH1 might influence the fusion of facial processes when the orofacial region is developing to its normal form 15,16. Metzis V et al showed that deletion of PTCH1 in the mouse embryos alters cell morphology and causes defective nasal pit epithelium invagination and cleft lip 17. Furthermore, PTCH1 mutations can cause Gorlin–Goltz Syndrome, in which orofacial clefts are frequently observed. These findings indicate the essential role of PTCH1 in forming calvaria and bone homeostasis 18,19.
RAD54B is a protein that in humans is encoded by the RAD54B gene. This protein plays a vital role in the DNA damage repair 20. The human RAD54B protein has been associated with NSCL/P 13,14. The results of the GWAS done by X Liu et al suggested that the rs12681366 in RAD54B could decrease the risk of NSCL/P in a Northern Chinese population 13. Qiao W et al found that the knockdown of RAD54B increased the sensitivity of Primary Mouse Embryonic Palatal Mesenchymal cells (MEPMs) to DNA double-strand break inducers 21. On the other hand, overexpression of RAD54B could increase cell apoptosis and diminish cell proliferation 22. Overall, these reports propose that RAD54B could have an essential regulatory role in NSCL/P incidence; although previous studies have found an association between PTCH1 and RAD54B with NSCL/P in Irish, Africans, northern, and southern Chinese. It is necessary to figure out if these associations can also be detected in different populations. Therefore, the purpose of the present retrospective case-control study was to evaluate the relationship between the rs10512248 and rs12681366 with NSCL/P in the northeast of Iran.
Materials and Methods :
Research subjects: This analytic cross-sectional study consisted of 120 cases with NSCL/P and 160 healthy subjects as control group. The study population was recruited between 2019-2020 from the Khayyam Hospital of Neishabur, Khorasan Razavi province, a city in the Northeast of Iran, in which previous reports have shown a high rate of NSCL/P. To exclude the syndromic forms of cleft lip/palate, oral and maxillofacial surgeons and medical geneticists precisely examined all cases to confirm that the cleft lip/palate is the only affecting disorder. The written consent and a questionnaire consisting of demographic information, family background of the specific disease, and parent's pregnancy history were also collected from all the subjects or their parents for the patients below 18 years old. This process was mandatory to rule out any probable prenatal contributory teratogenic factors that could have led patients to cleft lip/palate development.
The inclusion criteria for the control group subjects were as follows: (a) no cleft lip/palate or other inborn deformities, (b) no family history of congenital malformations, (c) no significant difference with the study group regarding age and sex, (d) no history of multiple miscarriages, cerebrovascular stroke, deep vein thrombosis, and cardiovascular diseases in the mother. In the case group, those NSCL/P patients with a mother's history of lack of iron or folic acid consumption during the pregnancy were excluded from the study.
Ethical statement: The protocols of this study were approved by the Research Ethics Committee of the Dental Faculty of Shahid Beheshti University of Medical Sciences with the code number IR.SBMU.DRC.REC.1399.027.
Sample collection: Blood samples were collected from all participants of both groups, or if not adults, by parents or legal guardians. Approximately 3 ml of peripheral venous blood was obtained from each subject. In order to prevent the blood samples from clotting, they were collected into EDTA-contained tubes (200 µl 0.5-M EDTA) and stored at -80°C until DNA extraction. DNA was immediately extracted from the leukocytes of blood using the standard salting-out procedures 23. The quantity and quality of extracted DNA samples were evaluated with the spectrophotometry and loaded in agarose gel, respectively. The concentration of DNA was assessed by measuring the Optical Density (OD) at the absorbance of 260/280 nm. Those samples whose absorbance ratio at 260 nm to 280 nm was 1.7-1.9 were considered pure DNA.
Genotyping: Genotyping of PTCH1 (rs10512248) and RAD54B (rs12681366) polymorphisms was determined using the Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP). The primer sequences are shown in table 1. Briefly, PCR was carried out into a 0.5 ml PCR microtube in a final volume of 25 µl comprising 5 μmol of each primer, 1 μl of dNTPs mix (Bioron, GmbH®, Germany), 30 ng of template DNA, 2.5 µl of 10×PCR buffer (Bioron, GmbH®, Germany), 1.5 mM MgCl2, and 0.5 U of Taq DNA polymerase (CinnaGen, Iran).
The cycling conditions for PTCH1 and RAD54B began with a denaturation step at 95°C for 4 min, followed by 33 cycles of 45 s of denaturation at 94°C, annealing temperature for 30 s at 60°C, and 40 s at 72°C, with a terminal elongation step of 5 min at 72°C. Subsequently, the cycles ended with a chilling phase to 4°C. Restriction digestion of PCR products was performed using Eco571(PTCH1) and NlaIII (RAD54B) enzymes at 37°C overnight. Electrophoresis on 2% agarose gels was used in order to separate DNA fragments (Figures 1 and 2).
Statistical analysis: All statistical analyses of the data were accomplished using SPSS software version 18.0 (SPSS Inc, Chicago IL, USA). A standard chi-square test (χ2) was performed to compare the frequencies of genotypes and alleles between the case and control groups. The p<0.05 was considered statistically significant. Odds ratios with 95% confidence intervals by unconditional logistic regression analyses were also utilized to estimate the relative relation between PTCH1 and RAD54B polymorphisms and the NSCL/P. Also, the independent sample t-test was used to compare the quantitative variables between groups. p<0.05 was considered statistically significant.
Results :
A total of 122 NSCL/P subjects (71 males, 51 females) and 161 healthy controls (94 males, 67 females) were recruited for our study. All patients were from Neyshabur city. The distributions of genotypes among the NSCL/P cases and controls revealed that females and males in case and control groups were in Hardy-Weinberg equilibrium. There were no statistically significant differences between the two groups regarding age and sex (p=0.975). According to the independent t-test results, case and control groups had no significant differences in mean maternal age (26.92±6.12 and 26.84±6.32 yrs, respectively) and participants' age (19.71±9.22 and 18.94±9.182 yrs, respectively). The distribution of genotypes and allele frequencies of the PTCH1 and RAD54B Polymorphisms in the affected subjects and controls are presented in tables 2 and 3.
The obtained results showed no significant difference between NSCL/P patients and the control group regarding PTCH1 (rs10512248) and RAD54B (rs12681366) polymorphisms in allele and genotype frequencies. In rs10512248, C allele frequency was higher in the control group (17.1%) compared with the case group (15.2%). According to univariate logistic regression analysis, CC and AA genotypes in the case group were higher than the control group. So, the AC genotype could correlate to a lower incidence of NSCL/P in the population than the AA genotype, however, the results were not statistically significant (p=0.615).
As for RAD54B (rs12681366), the frequency of the T allele (69.3) compared with C (30.7) was higher within the case group. Also, the C allele was higher in the control group (32.3%) compared with the case group (30.7%). About the genotypes, in the case group, the frequency of homozygote genotypes (TT, CC) was higher than in the control group. However, the heterozygote genotype (TC) of the case group (45.1%) was lower than the control (53.4%). These results were not significant (p=0.334).
Discussion :
Although the cleft lip and palate are among the most common congenital malformation in the craniofacial region, their prevalence differs among various geographical origins and ethnic groups 24. Both genetics and environmental factors are essential for assuming the main risk factors of NSCL/P. Previous studies have noted the association between BMP4 (rs17563), RFC1 (rs1051266), MTHFR (C677T), MSX1 (rs12532), FGF1 (rs34010), CDH1 (rs16260), and DHFR 19-bp insertion/deletion polymorphisms with NSCL/P in the southeast of Iran 25-30. The current study was the first investigation of the association between PTCH1 (rs10512248) and RAD54B (rs12681366) in the northeast Iranian population with NSCL/P. The results of the previous studies on the Irish, northern, and southern Chinese populations revealed the potential association between these two SNPs and NSCL/P 11-14. According to our results, no significant differences were found between the genetic polymorphism of rs10512248 and rs12681366 with NSCL/P in our population.
PTCH1 is a receptor for sonic hedgehog, a signaling molecule with an essential regulatory role in craniofacial morphogenesis, including palatal or labial clefts 31. In our study, the AA and CC genotypes frequency of PTCH1 was higher in the case group (74.6%-4.9%) than in the control group (70.2%-4.3%). On the other hand, the heterozygote genotype (AC) had more frequency in the control group (25.5%) than in the case (20.5%). Although these findings were not statistically significant in the present study (p=0.615), our results are in line with the research of Liu X et al 13. Their study suggested that the rs10512248 AC genotype could decrease the risk of NSCL/P (p=0.020) compared to the AA genotype in the northern Chinese population. However, it needs to be stated that after applying the Bonferroni correction in the mentioned study, the AC genotype did not remain a significant risk factor for NSCL/P. When evaluating differences between case-control and phenotypes, several hypothetical tests are often arranged. Multiple comparison tests can lead to a misunderstanding. Even in the absence of a significant result, there is a 64% chance of seeing a significant result, even if the tests themselves are insignificant. One method of reducing the chances of witnessing a significant result (p<0.05) is Bonferroni Correction 32.
RAD54B, an essential DNA damage repair protein, is located on 8p22.1 and belongs to the SWI2/SNF2 helicase superfamily, which modulates the DNA damage checkpoint response and homologous recombination repair pathway 33,34. Abnormal expression of RAD54B has been shown to be related to different types of cancers, such as colorectal cancer, lymphoma 35, and lung adenocarcinoma 36. Therefore, understanding the probable effect of RAD54B polymorphism on NSCL/P is critical. For rs12681366, the alleles and genotypes frequencies in the case and control subjects were not significantly different in the present study. The frequencies of the TT and CC genotypes among the cases (46.7%-8.2%) were higher than the controls (41.0%-5.6%). Also, the control group's CT genotype (53.4%) had more frequency than the case group (45.1%). In the Liu X et al study, the CT genotype in controls (54%) was significantly higher (p=0.001) than in cases (46%). This result could suggest that the rs12681366 polymorphism might decrease NSCL/P risk in the northern Chinese population. Though, this cannot be applicable to the Iranians, based on our study.
The difference observed in our results compared to Liu X et al 13 may be due to the different sample sizes (122 patients and 161 healthy members in the present study vs. 596 patients and 466 healthy members) and possible environmental and geographical diversity. Because as mentioned before, NSCL/P is a multifactorial disorder which means that besides the several gene polymorphisms, environmental factors can also alter the risk of incidence 4. Furthermore, previous studies have shown that the same genetic markers in different ethnic groups do not necessarily lead to the same phenotypic results in the head and neck 37,38. Therefore, the difference seen in the Chinese and Iranian populations in the PTCH1 (rs10512248) and RAD54B (rs12681366) can suggest that PTCH1 and RAD54B may be a multiethnic marker in NSCL/P.
Our current study has some limitations. First, our study's sample size could be a reason why our results were not significantly different in our population. Second, only one variant of the PTCH1 (rs10512248) and RAD54B (rs12681366) was evaluated in this study, other variants of PTCH1 and RAD54B should be considered for further assessment. Third, we assessed only one region and one ethnic group of patients, so the effect of ethnicity could not be evaluated. Future multicenter research with larger samples from different ethnicities is required.
Conclusion :
In conclusion, we performed this study to assess the probable association of PTCH1 and RAD54B gene polymorphisms with NSCL/P in the northeast Iranian population. Our results did not support this hypothesis. Although the genetic component of NSCL/P etiology has received widespread attention in recent years, most genetic variants contributing to this congenital craniofacial malformation are yet to be discovered. Our study is the first report evaluating the relation between rs10512248 and rs12681366 with NSCL/P in our population. Further research with larger sample sizes and different methods such as genome-wide association studies should be considered.
Acknowledgement :
This study was part of a thesis that has been supported by the Dentofacial Deformities Research Center, Research Institute of Dental Sciences, Shahid Beheshti University of Medical Sciences, Tehran, Iran. Their financial support is therefore highly appreciated.
Ethical approval :
The survey was approved by the Ethics Committee of the Dental Faculty of Shahid Beheshti University of Medical Sciences (Code No. IR.SBMU.DRC.REC.1399.027). The study was also preceded by completing written consent forms by all participants or their parents.
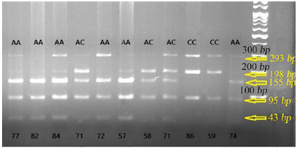
Figure 1. Photograph of RFLP method of PTCH1 (rs10512248) polymorphism: after digestion of PCR products with the restriction enzyme Eco571 for three genotypes of NSCL/P cases, one specific band of 155 bp was indicated in AA genotype, two specific bands of 198 and 155 bp were revealed in the AC genotype, and one specific band of 198 bp was indicated in CC genotype.
|
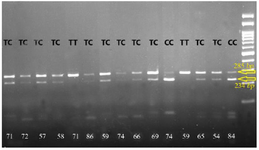
Figure 2. Photograph of RFLP method of RAD54B (rs12681366) polymorphism. After digestion of PCR products with the restriction enzyme NlaIII for three genotypes of NSCL/P cases, one specific band of 285 bp was indicated in TT genotype, two specific bands of 285 and 234 bp were revealed in the TC genotype, and one specific band of 234 bp was indicated in CC genotype.
|

Table 1. Primer sequences and their related sizes for each polymorphism
|
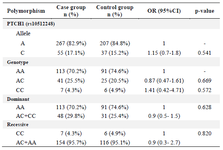
Table 2. Allele frequency and genotype distribution of the PTCH1 polymorphisms in case and control groups
|
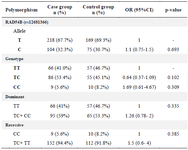
Table 3. Allele frequency and genotype distribution of the RAD54B polymorphisms in case and control groups
|
|