Antibodies Produced Toward Recombinant RBD and Nucleocapsid Neutralize SARS-COV-2
-
Rezaei, Amir
-
Department of Biology, Shahed University, Tehran, Iran
-
 Nazarian, Shahram
Molecular Biotechnology Research Center and Department of Biology, Imam Hussein University, Tehran, Iran, Tel: +98 21 51212232; Fax: +98 21 51212232; E-mail: nazarian56@gmail.com
Nazarian, Shahram
Molecular Biotechnology Research Center and Department of Biology, Imam Hussein University, Tehran, Iran, Tel: +98 21 51212232; Fax: +98 21 51212232; E-mail: nazarian56@gmail.com
-
 Mousavi Gargari, Seyed Latif
Department of Biology, Shahed University, Tehran, Iran, Tel: +98 21 51212232; Fax: +98 21 51212232; E-mail: slmousavi@shahed.ac.ir
Mousavi Gargari, Seyed Latif
Department of Biology, Shahed University, Tehran, Iran, Tel: +98 21 51212232; Fax: +98 21 51212232; E-mail: slmousavi@shahed.ac.ir
Abstract: Background: The highly contagious SARS-COV-2 virus spread rapidly from China and formed a global pandemic. The virus has infected over 509 million people worldwide and killed about 6.32 million up to date. Up on invasion, the Receptor Binding Domain (RBD) of Spike protein plays a crucial role in the entry of the virus into the host cell. The virus N protein is another protein that has a critical role for genome packaging.
Methods: As bioinformatics approaches, the cassette design, codon adaptation, and protein stability were investigated in this study. Synthetic genes of RBD and N were cloned separately in pET28a + expression vector. They were transferred into Escherichia coli (E. coli) BL21 DE3 host cell, and expression of recombinant proteins was induced with IPTG. The recombinant proteins were purified by column chromatography and approved by Western blotting. Animal immunization was performed with each of the recombinant proteins individually and in combination of the two. The antibody titer of the blood serum from control and immunized mice groups was determined by ELISA technique. Finally, the anti-spike neutralization test was performed.
Results: The expression and purification of RBD protein were monitored on SDS-PAGE, two bands of about 28 and 45 kDa for RBD and N appeared on gel distinctly, which were further validated by Western blotting. According to ELISA results, related antibodies were traced to a dilution of 1/64000 in immunized sera. The neutralization test exhibited produced antibodies' potency to bind the virus proteins. Using SPSS software, statistical analysis was performed by Duncan's test and T-test.
Conclusion: According to the present study, recombinant proteins, either RBD alone or in combination with N adequately stimulated the immune response, and the raised antibodies could neutralize the virus in in vitro test.
Introduction :
In 1960, the first human coronaviruses were discovered during research into cold-causing viruses in the United States and Britain 1,2. On December 29, 2019, hospital doctors in Wuhan, China, noticed unusual cases of patients with pneumonia 3. The ongoing coronavirus disease 2019 (COVID-19) pandemic became a serious public health crisis, caused by Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-2) 4. The primary host for the virus appears to be bats, however other organisms like pigs and snakes, can serve as a host 5,6. To date, many research groups have tried to find effective ways to fight the virus 7. According to the World Health Organization (WHO) report, until August 2021, the number of infected people was about 200 million and the number of dead was about 2.4 million (https://covid19.who.int); while the number of infected and dead population is increasing daily. Accordingly, developing efficient vaccines against the virus was considered as a priority, yet.
Coronavirus is a spherical virus that looks like a crown due to the large and small proteins on its surface 2,8. It is a small RNA-positive virus with a length of 29736 nucleotides; its genome has 79.5% similarity with SARS virus. The genome encodes four main structural proteins of the virus: Spike (S), Envelope (E), Membrane (M), and Nucleocapsid (N). Among them, Spike is more crucial and prepares the virus for invadind and attaching to the host cell 9,10. The N protein with 419 amino acids has more expression compared to other viral proteins and its immunogenic antibodies have been observed in infected patients 11-13. This protein surrounds the virus genome, and following the virus entrance in to host cell, the protein is separated and RNA is released into the cytoplasm 14. The N protein also acts as a chaperone and is essential for viral assembly 15,16. On average, there are 74 spike proteins on the surface of coronavirus 17. It divides into two parts; one part is responsible for attachment to the host cell and entrance. Another part is related to the placement and attachment to the virus membrane. The new coronavirus (COVID-19) uses the Angiotensin-Converting Enzyme 2 (ACE2) receptor to enter the host cell similar to that of SARS virus 18. The virus enters through the Receptor-Binding Domain (RBD) in the S1 subunit. The SPIKE protein detects Lys 31 in ACE2 through Gln 394 in the RBD domain 18,19. After binding to the ACE2, the spike is cleaved by acid-dependent proteolysis of Human Airway Trypsin-like protease (HAT) and cathepsin and transmembrane protease Ser 2 (TMPRSS2), into two subunits of S1 and S2 20. The S1 fragment consists the RBD section and the S2 fragment contains the C-terminal with the HR1 and HR2 (Heptad Repeat) sequences 21,22. The RBD is the target of many primary neutralizing antibodies, for preventing the virus from binding to the receptor (ACE2/DPP4). Therefore, RBD of spike can be a proper candidate for vaccine development against coronavirus. It has been reported that co-administration of protein subunits increases the immunogenicity compared to single administrations 23. Therefore, in the present study, RBD and N proteins were expressed in Escherichia coli (E. coli), and purified discretely. The recombinant RBD, alone or in RBD-N combinations were compared as immunogenic candidates against SARS-COVID-2 in mice model. The antibodies produced were used in virus neutralization tests.
Materials and Methods :
In silico design of the construct: Bioinformatics tools have a great impact on the design analysis of immunogens. RBD and N protein sequences with accession numbers NC_045512.2 and sp-P0DTC9 were obtained from the GeneBank and UNIPROT databases in bioinformatics approaches. The sequences were aligned through the BLAST tools at the GeneBank site for confidence. PROTPARAM tools of EXPASY analyzed structural and physical information of the proteins, and the I-TASSER software was utilized to predict the three dimensional structure of proteins. The Ramachandran plot tool in the RAMPAGE server was utilized to confirm protein structures by measuring the statistical distribution of the combinations of the backbone dihedral angles ϕ and ψ. BCEpred online software was employed to predict the induction of humoral immunity, and VaxiJen online software was used to estimate the overall antigenicity of the sequences.
Expression and purification of the recombinant proteins: The synthetic genes were obtained on pUC18 and subcloned into the pET28a vector with Nedl and BamHl restriction sites at Imam Hussein University, Tehran, Iran. The constructs were transformed into E. coli Bl21 (DE3) and cultured on an LB agar plate containing 70 µg/mL kanamycin.
Cells were induced with 1 mM Isopropyl β-d-1- thiogalactopyranoside (IPTG) for five hours. Cells were harvested and lysed with sonication. The recombinant proteins as inclusion bodies were dissolved in phosphate buffer (100 mM NaH2PO4, 10 mM Tris-HCl) containing 8 M urea and were analyzed on 15% SDS-PAGE.
Recombinant proteins were purified using Nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography. Each column was equilibrated with phosphate buffer and loaded with approximately 500 µg of protein. The column for protein N was washed five times with a 20 mM concentration of imidazole, and the recombinant protein was eluted with a 250 mM concentration of imidazole. To purify the RBD protein, the column was washed seven times with phosphate buffers: pH=6.3 (C buffer), pH=5.5 (D buffer), and pH=4 (E buffer). The recombinant protein was eluted with 250 µl of E buffer. All collected fractions were analyzed on the SDS-PAGE, and concentration proteins was assessed with Bradford method.
Western blot analysis: The purified recombinant proteins were transformed onto the nitrocellulose membrane with transfer buffer (39 mM glycine, 48 mM Tris-base, 0.037% SDS, and 20% methanol). The membrane was blocked with blocking buffer (PBST+5% Skimmed Milk Powder) for 15 hr at 4°C and washed three times with PBST (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4+0.05% Tween). Nitrocellulose membranes were soaked in mice anti his-tag IgG (1:10000), and incubated for one hour at 37°C. Membranes were washed and added with DAB (3, 3´-diaminobenzidine) as a chromogenic substrate. The reaction was stopped by distilled water.
Animal immunization: Twenty-four female BALB/c mice (20-25 gr, 5-week old) were obtained from Tehran University, Tehran, Iran. The mice were divided into four groups. Two groups each with nine mice for immunization, were named as RBD and RBD-N groups. Two other groups each with three mice were kept as control groups. Mice were kept in the animal care facility of Shahed University under standard and ventilated conditions. The principles in the guide for the care and use of laboratory animals were followed. All animals were kept in compliance with the Welfare act (Helsinki and its later amendments). The animal care rule was ethically certified by Shahed University with the ethical code of IR.SHAHED.REC.1400.018.
Mice in RBD-N group were subcutaneously injected with 100 µl of recombinant proteins mixed with equal volume of Allum adjuvant on three boosters on days 0, 15, and 30. The control group received Phosphate-Buffered Saline (PBS) with Allum adjuvant (Table 1). Blood samples were collected from the eye corners of mice on days 14, 29, and 44 after the first injection. The sera were collected and stored at -20°C.
Antibody response: Enzyme-linked Immunosorbent Assay (ELISA) technique was applied to determine antibody response. 5 µg of each recombinant protein (in 100 µl of coating buffer) was coated in ELISA wells and incubated at 4°C overnight. The wells were blocked with skimmed milk, and then the serial dilution (1:100 to 1:64000 in PBST) of serum antibodies was added to the wells and incubated at 37°C for 2 hr. Anti-mouse IgG HRP conjugate (1/2000 dilution in PBST) was added to each well and incubated at 37°C for 90 min. 100 µl Tetramethylbenzidine (TMB) was added to each well and incubated at room temperature for 10 min. Wells were washed after each incubation. The reactions were stopped with 3N H2SO4, and the OD450 was recorded.
Neutralization tests: The neutralization of the SARS-CoV-2 was performed using a laboratory kit supplied by PishtazTeb Zaman Diagnostics. This kit contains the virus spike protein and ACE2 receptors. The ACE2 receptors in the kit will not be able to bind to the spike proteins in the presence of anti-RBD antibodies. Tests were performed according to the protocol provided by the supplier.
Statistical analysis: The data in each figure was a representative of three independent experiments expressed as the mean± standard deviation (SD). All statistical analyses were performed using a SPSS 12.0 statistical program. Student t-test was used to analyze the data for antibody responses between immunized and non-immunized groups. A value of p<0.05 was considered statistically significant.
Results :
Bioinformatics analysis: Validated sequences were obtained from the GenBank database. The three-dimensional structures of proteins were predicted by the I-TASSER software (Figure 1). According to this tool's description, a Confidence score (C-score) is utilized for estimating the quality of predicted models by the server. Its algorithm is based on the significance of threading template alignments and the convergence parameters of the structure assembly simulations. C-score is typically in the range of 2-5, where a C-score of higher value signifies a model with high confidence and vice-versa. RBD protein structure was predicted with a C-score of 1.05, and for the N protein, it was -3.56. The Ramachandran plot quality assessment analysis showed that most RBD protein residues, i.e., more than 98%, are in the favored + allowed region. Most N protein residues, i.e., more than 99%, are in the favored area as well and hence these structures can be reliable. The B cell epitopes were also predicted as shown in tables 2 and 3.
Expression and purification of the recombinant proteins: Expression and purification of recombinant RBD (28 kDa) and N (45 kDa) proteins was analysed on SDS-PAGE. The SDS-PAGE and Western blotting method verified the recombinant proteins (Figures 2-5).
Determination of serum IgG titer: Indirect-ELISA was performed to evaluate the raised IgG antibody titers of sera. After second blood sampling, antibody titers measurement exhibited a significant increase in the test groups compared to the control ones. High antibody titers were observed in both experimental groups until day 87 following the first injection. The RBD-N group developed more robust and more persistent immunogenicity than the RBD alone (Figure 6).
Neutralization tests: According to the supplier's instructions, if the average concentration of antibodies from the vaccine is more than 2.5 µg/ml, the neutralization test is considered positive. The mean light absorption of antibodies from the RBD single vaccine in virus neutralization was 0.6186, corresponding to 19.48 µg/ml of RBD antigen alone. The mean light absorption of antibodies from the combination of RBD-N antigens in virus neutralization was 0.3949, corresponding to 25.17 µg/ml of RBD-N mixture.
Discussion :
Since June 2020, about 157 vaccine candidates have been tested for the COVID-19 in laboratories worldwide. Several vaccines, including inactivated viruses, adenovirus vectors containing the spike protein, and mRNA vaccines against the virus have been used in clinical and commercial development 24-26. Because of their safety concerns and high efficacy, about 53% of researches have focused on recombinant vaccines 27. Available research data from the Mers virus indicates that inactivated viruses and recombinant vaccines efficiently neutralize viruse activity 28. Today, inactivated viruses are produced by UV and formaldehyde treatment. However, viruses such as corona cannot be cultivated at any laboratory and require a level three laboratory, which is challenging to establish and maintain due to the high cost. At high-level production of attenuated virus, checking each batch's inactivation is another problem concerned with these vaccines 29,30. Furthermore, inactive microbes cannot multiply in the host, and they must be used with adjuvants and administered in multiple doses of the vaccine 31. Some antibodies are produced against parts of the pathogen but having no role in causing the disease. In this case, cytotoxic T cells are not produced and can not be used to stop infections by intracellular pathogens 32. Animals vaccinated with inactivated bacteria developed fever at the end of the experiment, whereas animals immunized with the recombinant protein showed no such side effects 33. S1, S2, and RBD were the first recombinant vaccine candidates, of which RBD remains an essential candidate for primary vaccines and boosters due to the high neutralizing effect and CD4+ T cell epitopes 34-37. Interestingly, the RBD effectively boost immune responses against emerging variants 38. The RBD domain is the most concentrated area of epitopes in most articles and has been regarded as an attractive immunogen 39-41. In contrast, the S1 protein induced a lower neutralization titer than the RBD protein 42 and hence the RBD is a better candidate than S1 for SARS-CoV-2 vaccine design. The toxicity of recombinant RBD protein is very low and has no effect on weight, appetite, and behavior. Also, it has no adverse pathological effects on the brain, liver, kidneys, or heart and does not reduce the number of blood cells 42. In these experiments, aluminum adjuvants were selected as one of the best immunogenic adjuvants, and unlike Freund's adjuvant, this adjuvant can also be used for humans 42,43.
Recombinant RBD and N proteins were expressed in the bacterial host at approximately 40 mg/L for RBD and 50 mg/L for N protein. The average expression of RBD and N in eukaryotic hosts are 45 mg/L and 50 mg/L, respectively 44-48. Despite the glycosylation in eukaryotic cells and the sugar removal process, it seems appropriate for expressing these proteins in the bacterial host. In addition, these proteins maintain their stability at -70°C and can be stored for a long time.
In our study, the average percentage of neutralization by the serum induced toward RBD alone was 48.7%, and increased to about 63% with serum of mice immunized both with RBD and N proteins. The increased percentage of neutralization in the presence of N can be attributed to the adjuvant role of N rather than its role as an immunogen. In a similar work, antibodies from the RBD vaccine prevented the protein from binding to its receptor by 20 to 40% 49.
Conclusion :
Combination vaccines are a new way to make vaccines safer, more durable, and more effective. Recombinant measles viruses containing proteins S and N on their surface provided better immunity 50. The immunogenicity of the single RBD vaccine has been compared with the RBD-dimer. The RBD monomer acts as a poor immunogen and triggers low levels of IgG and neutralization results 51. Data presented here and the results obtained by other researchers, shows that RBD as a recombinant vaccine candidate, with its simple, rapid, and economical preparation, might be an effective and convenient strategy to control COVID-19 epidemics.
Acknowledgement :
The authors are thankful to Shahed University and Imam Hussein University for supporting this work.
Conflict of Interest :
The authors declare no conflict of interest.
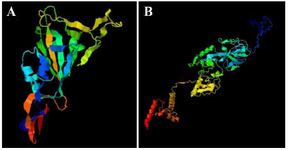
Figure 1. A) Three-dimensional structure of RBD. B) three-dimen-sional structure of N protein.
|
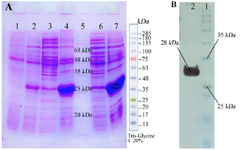
Figure 2. A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of RBD expressions: Lane 1: RBD clone uninduced with IPTG as control extract in B buffer, Lane 2: RBD clone induced with 0/5 mM IPTG, 4 hr and extract in B buffer, Lane 3: RBD clone induced with 1 mM IPTG, 4 hr and extract in PBS buffer, Lane 4: RBD clone induced with 1 mM IPTG, 4 hr and extract in B buffer, Lane 5: Molecular weight marker, L ane 6: RBD clone induced with 0/5 mM IPTG, over night and extract in B buffer, Lane 7: RBD clone induced with 1 mM IPTG, over night and extract in B buffer. B) Western Blot analysis of the RBD protein, Lane 1: Molecular weight marker, Lane 2: RBD protein.
|
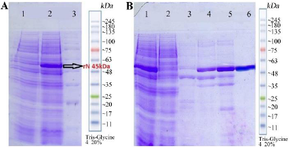
Figure 3. A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of N expressions: lane1: N clone uninduced with IPTG as control. Lane 2: N clone induced with Isopropyl β-d-1-thiogalactopyranoside (IPTG), lane 3: Molecular weight marker. B) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of N purification: lane 1: Pre Flu, lane 2: Flu, lane 3: I40 (1 ml), lane 4: I100 (1 ml), lane 5: I150 (1 ml), lane 6: I250 (500 μl).
|
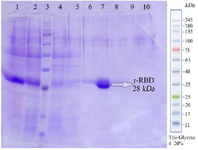
Figure 4. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of RBD purification. Lane 1: sample before purification, lane 2: Flu, lane 3: Molecular weight marker, lane 4: C(2 ml), lane 5: D(2 ml), lane 6: E1(500 μl), lane 7: E2(500 μl), lane 8: E3(500 μl), lane 9: E4(500 μl), lane 10: Cu Buffer(1 ml).
|
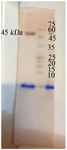
Figure 5. Western Blot analysis of the N protein, Lane 1: Molecular weight marker, lane 2: N protein.
|
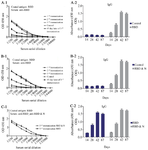
Figure 6. Linear and bar charts compare ELISA antibody titers in three blood samples in safe and control mice. A-1) Line chart of ELISA antibody titer obtained from single RBD vaccine in three blood samples 2 weeks apart and final blood sample 45 days after the last immunization. A-2) Column chart of ELISA antibody titer obtained from single RBD vaccine in three blood samples 2 weeks apart and final blood sample 45 days after the last immunization. B-1) Line chart of ELISA antibody titer from RBD & N combination vaccine in three blood samples 2 weeks apart and final blood sample 45 days after the last immunization. B-2) Column chart of ELISA antibody titer from RBD & N combination vaccine in three blood samples 2 weeks apart and final blood sample 45 days after the last immunization. C-1) Comparison of linear graphs of ELISA antibody titers from RBD single vaccine and RBD & N combination vaccine in the third blood sample. C-2) Comparison of Column graphs of ELISA antibody titers from RBD single vaccine and RBD & N combination vaccine in the third blood sample.
|

Table 1. Mouse immunization protocol
|
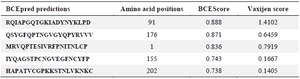
Table 2. B-cell epitopes from RBD protein; using BCEPred
Antigenicity of RBD protein; using VaxiJen
|
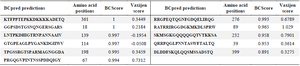
Table 3. B-cell epitopes from N protein; using BCPred
Antigenicity of N protein; using VaxiJen.
|
|