Association of MTHFR, BMP4, TGFA and IRF6 Polymorphisms with Non-Syndromic Cleft lip and Palate in North Indian Patients
-
Avasthi, Kapil
-
Department of Medical Genetics, Sanjay Gandhi Postgraduate Institute of Medical Sciences (SGPGIMS), Lucknow 226014, India
-
Agarwal, Amit
-
Department of Burn and Plastic Surgery, Vivekananda Polyclinic and Institute of Medical Sciences (VPIMS), Lucknow 226007, India
-
 Agarwal, Sarita
Department of Medical Genetics, Sanjay Gandhi Postgraduate Institute of Medical Sciences, saritasgpgi@gmail.com
Agarwal, Sarita
Department of Medical Genetics, Sanjay Gandhi Postgraduate Institute of Medical Sciences, saritasgpgi@gmail.com
-
Department of Medical Genetics, Sanjay Gandhi Postgraduate Institute of Medical Sciences (SGPGIMS), Lucknow 226014, India
Abstract: Background: Non-Syndromic Cleft Lip and Palate (NSCL/P) is a multifactorial birth defect. The world-wide prevalence of NSCL/P is 1 in 1000 live births; it differs with race, ethnicity and gender. The aim of the present study was to find out the status of candidate gene polymorphisms in NSCL/P cases and its association in phenotype of the patients.
Methods: We have screened five polymorphisms in four candidate genes MTHFR (rs1801133, rs1801131) BMP4 (rs17563), TGFA (rs1146297) and IRF6 (rs2235371) by restriction fragment length polymorphism and results were validated by Sanger sequencing. Our dataset consists of 200 NSCL/P cases and 200 healthy controls from the Indian population. Statistical data analysis was performed by SPSS software.
Results: MTHFR (rs1801133), BMP4 (rs175563) and TGFA (rs11466297) gene polymorphisms showed significant association with NSCL/P and act as a risk factor in the Indian population (p=<0.05). However, MTHFR (rs1801131), and IRF6 (rs2235371) gene polymorphisms did not show significant association with NSCL/P in the Indian population.
Conclusion: The result of the study suggests an association between MTHFR (rs1801133), BMP4 (rs175563) and TGFA (rs11466297) polymorphisms with NSCL/P in Indian population.
Introduction :
Non-Syndromic Cleft Lip and Palate (NSCL/P) is the world's second most prevalent congenital birth defect, with incidence of 1 in 700 live births and varies by ethnicity or geographical region. Balaji et al, reported a prevalence of NSCL/P 1.3 in 1000 live births in India 1. Since patients of NSCL/P suffer with problems of feeding, speech difficulties, malnutrition, hearing injuries, infections and mental disorders from birth to adulthood, they need multidisciplinary care like surgical or dental treatment, speech therapy and psychosocial interventions throughout life 2. A sequence of closely orchestrated events are needed during development of lip and palate formation, including cell proliferation, growth, differentiation and apoptosis. The disruptions in any of these events affects unacceptable facial structure morphology resulting in manifestation of disease 3,4. Thus it is considered as a complicated hereditary disorder but polygenic in nature. Shi et al, reported involvement of environmental risk factors like tobacco, smoke and alcohol intake during early pregnancy 5.
Epidemiological surveys and animal model studies have also shown that antiepileptic medications or hormonal treatment, are the risk factor for NSCL/P 6. In past linkage analysis, association studies, direct sequencing, and more recently genome-wide association studies have been done in relation to NSCL/P and found suitable for genetic predisposition studies 2. Approximately 20 gene loci are identified in NSCL/P etiology, among those genes; we have selected five polymorphisms from four genes for present study, which are MTHFR (rs1801135, rs1801131), BMP4 (rs17563), TGFA (rs1146297) and IRF6 (rs2235371).
Methylenetetrahydrofolate reductase (MTHFR) is one of the most important enzymes which plays a crucial role in the folate metabolism regulation. The genecoding for MTHFR is on the long arm of chromosome 1 (1p36.3), which contains 11 exons 7. MTHFR (C677-Tand A1298C) has two common single nucleotide polymorphisms responsible for a moderately variable enzymatic action. MTHFR gene polymorphism studies have been reported from several regions suggesting a strong association with NSCL/P. Bone Morphogenetic Proteins (BMPs) plays an important role in the fusion of the upper lip, main palate, and craniofacial growth, primarily expressed in palatal shelf epithelial and mesenchymal cells 8-10. Saket et al, have suggested that the polymorphism of BMP4 (rs17563) variant plays a significant role in the frequency of CL/P in the Iranian population 11. The importance of BMP4 (rs17563) variations in the development of CL/P was addressed in previous investigations 9,10. Throughout craniofacial growth, TGFα is expressed in the inner epithelium boundary of fusing palatal shelves and activates the extracellular matrix synthesis 12,13. Ardinger et al, and Ebadifar et al, assessed the role of TGFA gene polymorphism in the event of CL/P and results show a strong association 14,15. Interferon Regulatory Factor 6 (IRF6) is a transcription factor and is located on chromosome 1q32.3-q41 16. IRF6 gene assembly consists of a highly conserved helix-turn-helix DNA-binding domain and a less conserved protein binding domain. The IRF6 gene is the gene between the candidates involved in both syndromic and non-syndromic form CL/P, with variations in this gene associated with van der Woude syndrome 17. The IRF6 gene polymorphism rs2235371 is well established in many studies of NSCL/P 18.
As the genetic diversity of NSCL/P is present in different ethnic groups, the role of these gene remains speculative in Indian populations; therefore, in this study, we have aim to investigate the association of genetic polymorphism of MTHFR, BMP4, TGFA, and IRF6 in NSCL/P in Indian population.
Materials and Methods :
Study design and ethical approval
This study was carried out from September 2017 to December 2020 in the Department of Medical Genetics, Sanjay Gandhi Postgraduate Institute of Medical Sciences, Lucknow. The research was carried out according to the World Medical Association (Declaration of Helsinki) for Experiments in humans. The study was approved by the Institutional Ethical Committee (IEC No: 2018-107-EMP-EXP) and informed consent was obtained from cases and controls.
Sample collection
NSCL/P patient’s sample (n=200, age=7±5 years) and age and sex matched healthy controls (n=200, age=7±5 years) were recruited for the study, after signed informed consent was obtained from patients and parents. The exclusion criteria for NSCL/P cases were patients with any other history of developmental disorder, Syndromic form of CL/P (e.g., eye, brain, limb anomalies and cardiac defects). The healthy controls were Indian healthy children without cleft lip and palate and other known genetic diseases.
Genotyping analysis
DNA isolation: Genomic DNA was extracted from 3 ml peripheral venous blood samples of patients and controls. DNA was extracted by using standard Phenol-chloroform method, quantification of the DNA was measured by spectrophotometer at wavelength of 260 nm and the quality was checked on 0.8% agarose gel.
Restriction fragment length polymorphism (RFLP)
Primers were designed to amplify candidate gene variants MTHFR (C677T, A1298C), BMP4 (T<C), and TGFA (A<C). Polymerase Chain Reaction (PCR) performed details of primers, annealing temperature and restriction sites are mentioned in table 1. The PCR amplified products were digested using restrictions enzymes (Hinfl for C677T, MboII for A1298C, Hphl for T<C and BamHI for A<C) and were kept at 37°C overnight; products were visualized by standard Ethidium Bromide-Agarose Gel Electrophoresis method (Figures 1-4).
Sanger sequencing
Sanger sequencing was done for the IRF6 (G<A) gene polymorphism followed by amplification and PCR product purification. Sequencing of PCR products were performed using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, California, United States), and products were resolved on the ABI 3130XL Genetic Analyser (Applied Biosystems). Sequence electro-pherograms were analysed using Finch TV (Figure 1-5).
Statistical analysis
Genotype and allele frequency distributions of MTHFR BMP4, TGFA and IRF6 polymorphisms were calculated by counting the genotypes and compared with the predicted values using the chi-square test, based on the assumption of Hardy-Weinberg equilibrium. The Odds Ratios (OR) were calculated with the 95% confidence intervals (95% CI) and p-values <0.05 were considered to be significant. All analyses were performed using SPSS for Windows, version 18.0 (SPSS Inc., Chicago, USA).
Results :
This study consisted of 200 NSCL/P patients (102 males, 98 females) and 200 healthy individuals (105 males, 95 females). The observed genotype frequencies of cases and controls in all polymorphic sites were in Hardy-Weinberg equilibrium. The genotyping results, OR (95% CI) and p-value calculations for five Single Nucleotide Polymorphisms (SNPs) of the MTHFR (rs1801133, rs1801131), TGFA (rs1146297), BMP4 (rs17563) and IRF6 (rs2235371) are reported in table 2 (Figures 1-5).
Findings revealed that MTHFR rs1801133 (OR= 1.56, 95% CI: 1.02-2.39, p=0.041), BMP4 rs17563 (OR=1.85, 95% CI: 1.19-2.89, p=0.005) and TGFA rs1146297 (OR=1.69, 95% CI: 1.01-2.82, p=0.045) polymorphisms are significantly associated with NSC-L/P. Our findings did not support an association between MTHFR rs7224837 and IRF6 rs861019 polymorphisms and risk/protection of NSCL/P (Table 2).
Discussion :
The aetiology of orofacial clefts remains largely unknown, although genetic factors are thought to play the most important roles. There is compelling evidence suggesting that common genetic variations contribute to NSCL/P susceptibility 2. In this study, we systematically evaluated the associations between a comprehendsive panel of five polymorphisms of the four genes MTHFR, BMP4, TGFA and IRF6 involved in NSCL/P. MTHFR C677T allele will increase the incidence of NSCL/P in Asian and Chinese populations 19,20. Furthermore, MTHFR 677TT homozygotes are associated with NSCL/P and 6777CT heterozygotes is the minor risk factor. However, a study from southern Han Chinese population reported involvement of MTHFR 677CT in cleft lip only 21. Studies from southern and northern part of India also reported association between MTHFR C677T and NSCL/P 22,23. Studies reported from China and Thailand showed no association between MTHFR C1298A allele risk factor for NSCL/P, which supports our results 24-27. Also MTHFR A1298C poses no risk of any combination with NSCL/P in the eastern Uttar Pradesh 23.
Several studies reported earlier have shown that BMP4 may be involved in CL/P formation. Hu, et al found in meta-analysis, depending on the ethnicity range, the BMP rs17563 variant plays a different role in NSCL/P. This variant significantly increased the risk of NSCL/P in the Chinese population, while the Brazilian population showed a protective effect 28,29. By contrast, Chen et al, reported that this variant was not associated with NSCL/P in the Asian population 30.
First report in NSCL/P with TGFA gene polymorphisms was reported by Ardinger et al, and shows significant association with TGFA rs11466297 14. Several studies have been published on TGFA. Studies of British, Japanese and French populations indicate involve-ment of the TGFA rs11466297 polymorphism in the occurrence of NSCL/P (31-33). Lidra et al, from Philippines published a contradictory study; it may be due to genetic differences in different populations 34. Past research findings are contradictory and may be due to variations in sample size, demographic history and environmental conditions. Ebadifar et al, found that there is a link between BamHI variant polymorphism and prevalence of CL/P in the Iranian community, so that the incidence of AC genotype and C allele in the patient sample was substantially higher 15.
Zucchero et al investigated 36 SNPs in IRF6 gene in 10 populations including Asian, European and American; among these IRF6 gene polymorphism rs2235371 was reported as a risk factor for NSCL/P in Filipinos of Asia 16. Association studies between IRF6 gene rs2235371 polymorphism and NSCL/P are well documented in Norway and western Chinese population 35,36. Rahimnov et al, reported a lack of involvement of polymorphism rs2235371 and NSCL/P in the Brazilian population 18. A study in south Indian population indicated IRF6 (rs2235375) gene polymorphism is significantly associated with increased risk of NSCL/P 37. Study from eastern part of Uttar Pradesh reported minor risk of IRF6 820GG with NSCL/P 23.
Conclusion :
The present study assessed the interaction effects of MTHFR (rs1801133, rs1801131), BMP4 (rs17563), TGFA (rs11466297), and IRF6 (rs2235371) polymorphisms on the occurrence of NSCL/P in Indian population. The results showed that the MTHFR (1801133), BMP4 (rs17563), and TGFA (rs11466297) polymorphisms have a significant effect on the occurrence of NSCL/P.
Acknowledgement :
The author would like to thank DST-INSPIRE, Ministry of Science and Technology, India Sanjay Gandhi Post Graduate Institute of Medical Sciences SGPGIMS, Lucknow, Uttar Pradesh and the Smile Train Foundation.
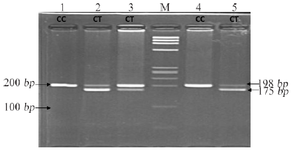
Figure 1. Restriction fragment length polymorphism result of MTHFR rs1801133 (C>T).
|
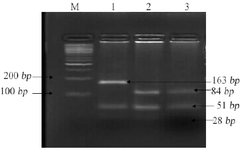
Figure 2. Restriction fragment length polymorphism result of MTHFR rs1801131 (A>C).
|
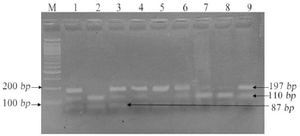
Figure 3. Restriction fragment length polymorphism result of BMP4 rs17563 (T>C).
|
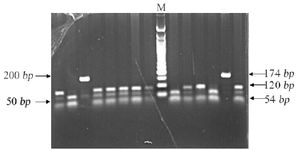
Figure 4. Restriction fragment length polymorphism result of TGFA rs11466297 (A>C).
|

Figure 5. Sanger Sequencing result of IRF6 rs2235371 (G>A).
|

Table 1. Candidate gene Variants Details
|
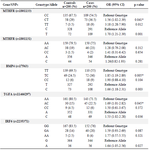
Table 2. The genotype and allele frequency of the MTHFR, BMP4, TGFA and IRF6 polymorphisms in the case and control groups
|
|