A Cross-Sectional Study for Evaluation of KRAS and BRAF Mutations by Reverse Dot Blot, PCR-RFLP, and Allele-Specific PCR Methods Among Patients with Colorectal Cancer
-
Sheikhsofla, Fatemeh
-
Department of Cellular and Molecular Biology, University of Mazandaran, Mazandaran, Iran
-
 Poopak, Behzad
Department of Clinical Laboratory Sciences, Islamic Azad University, Tehran Medical Sciences Branch, Tehran, Iran, Tel: +98 21 22264144; Fax: +98 21 22264144; E-mail: F.sheikhsofla@gmail.com
Poopak, Behzad
Department of Clinical Laboratory Sciences, Islamic Azad University, Tehran Medical Sciences Branch, Tehran, Iran, Tel: +98 21 22264144; Fax: +98 21 22264144; E-mail: F.sheikhsofla@gmail.com
-
Firuzyar, Sajjad
-
Razi Vaccine and Serum Research Institute of Karaj, Karaj, Iran
-
Ghadiany, Mojtaba
-
Department of Hematology and Oncology, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Abstract: Background: KRAS and BRAF genes are the biomarkers in Colorectal Cancer (CRC) which play prognostic and predictive roles in CRC treatment. Nowadays, the selection of rapid and available methods for studying KRAS and BRAF mutations in anti-EGFR therapy of patients suffering from CRC plays a significant role. In this study, the mutations of these two oncogenes were evaluated by different methods.
Methods: This study was performed on 50 Formalin-Fixed Paraffin-Embedded (FFPE) tissue blocks of patients diagnosed with colorectal cancer. After DNA extraction, KRAS and BRAF gene mutations were evaluated using reverse dot blot, and results were compared with PCR-RFLP and allele-specific PCR for KRAS and BRAF mutations, respectively.
Results: KRAS gene mutations were detected in 42% of patients, of which 30% were in codon 12 region, and 12% in codon 13. The most frequent mutations of KRAS were related to G12D and 10% of patients had BRAF mutated genes. The type of KRAS gene mutations could be evaluated by reverse dot blot method. In general, the results of PCR-RFLP and allele-specific PCR were similar to the findings by reverse dot blot method.
Conclusion: These findings suggest that PCR-RFLP and allele-specific PCR methods are suitable for screening the presence of the mutations in KRAS and BRAF oncogenes. In fact, another method with more sensitivity is needed for a more accurate assessment to determine the type of mutations. Due to higher speed of detection, reduced Turnaround Time (TAT), and possible role of some KRAS point mutations in overall survival, reverse dot blot analysis seems to be an optimal method.
Introduction :
Colorectal Cancer (CRC) is the third most common cancer in men, the second most common one in women, and the fourth most common one worldwide 1. Despite advancements in the diagnosis and treatment of CRC, approximately 35% of patients with CRC show stage IV or metastasis of the disease, and about 20 to 50% of patients show stage II and III of disease 2. The development of colorectal cancer is a multi-stage process that is associated with genetic changes 3. The transformation of a normal colonic epithelial cell into a cancer cell is characterized by the activation of oncogenes and the inactivation of tumor suppressor genes 4. Different molecular mechanisms are involved in the biological course of colorectal cancer development. In the genetic pathway, the conversion of normal cells to the proliferative epithelial cells is associated to the mutations in Adenomatous Polyposis Coli (APC) gene and the conversion of proliferative epithelial cells to an adenoma is also linked to changes in DNA methylation. At the adenoma stage, mutations in the BRAF and KRAS genes occur. Finally, adenoma transformation into carcinoma is associated with a mutation in the P53 gene 5. KRAS protein is a small GTPase which plays an important role in the transduction of intracellular signals. This gene is a member of MAP kinase pathway 6. The mutation in KRAS is capable of reducing this gene's protein activity and thus decreases GTPase activity 7. The prognostic and predictive role of KRAS was determined in literature by studying on this oncogene. It also encodes the cytoplasmic serine-threonine kinase, BRAF gene, which immediately activates MAPK signaling pathway downstream of KRAS. Abnormal activation of BRAF gene causes cell proliferation and survival which are important for tumorigenesis in many types of tumors 8. The role of KRAS and BRAF mutations in survival of CRC patients and response to standard chemotherapy are still controversial issues. Therefore, the study of these mutations is of great importance for patients with CRC. A study by Garcia-Carbonero et al on 792 CRC patients identified KRAS and BRAF genes as prognostic biomarkers as well as biomarkers of standard first-line chemotherapy response in metastatic colon cancer 9. In CRC management, detecting the absence of KRAS, BRAF, NRAS, and PIK3CA gene mutations with the choice of anti-EGFR therapy is one of the most useful strategies 10. Due to the high importance and role of these two oncogenes as CRC biomarkers and advancements in technology, different methods were developed to study and evaluate KRAS and BRAF mutations. Next-generation sequencing, real-time PCR, PCR-RFLP, allele-specific PCR (ASPCR), pyrosequencing, reverse dot blot, sequenom genotyping assays, LNA-PCR sequencing, HRM sequencing, multiplex mutation assay, and HRM-SNaPshot are a short list of different techniques for detecting KRAS and BRAF mutations. In 2016, Matsunaga et al compared the result of KRAS mutations by direct sequencing, Scorpion-ARMS assays, pyrosequencing, and Luminex xMAP methods 11. Nagakubo et al compared different methods (Sanger sequencing, PCR–reverse sequence-specific oligonucleotide probe, and next-generation sequencing) to detect KRAS, BRAF, and NRAS mutations 12.
Among these different methods, reverse dot blot, PCR-RFLP, and allele-specific PCR were used to perform this experiment. First, reverse dot blot was performed to detect KRAS and BRAF mutations and results were compared by PCR-RFLP and allele-specific PCR for KRAS and BRAF mutations, respectively. The mechanism of PCR-restriction fragment length polymorphism (PCR-RFLP) is based on discovering a specific recognition site in a target sequence 13. PCR-RFLP is a method which uses restriction enzymes to digest DNA, and the digested fragments will be loaded on the gel. In this method, if there are genetic changes or a mutation, it appears as a change in the size of one or two fragments in the gel electrophoresis due to insertion or deletion 14. Allele-specific PCR method has been used to detect Single Nucleotide Polymorphisms (SNPs) or bands of wild-type and mutant alleles, and the PCR product will be loaded on the gel. Reverse allele-specific oligonucleotide assay (Reverse dot blot) is a method for the molecular characterization of disorders with high mutation spectrum 15. In reverse dot blot hybridization, after a multiplex PCR, PCR products are hybridized to wild-type and mutant oligonucleotide probes on a nitrocellulose membrane 14, and then the positive and control bands will appear on a nitrocellulose paper.
Materials and Methods :
In this cross-sectional study, 50 FFPE tissue blocks belonging to patients with colorectal cancer with a mean age of 56.9 years (34 males and 16 females) were randomly collected from cases who referred to Payvand clinical laboratory from different parts of the country. Each patient completed a questionnaire about age, gender, city of residence, and treatment history, and before extraction, the blocks were checked by each patient's pathology report.
Genomic DNA extraction: To investigate the mutations of KRAS and BRAF genes, genomic DNA was extracted from FFPE tissue blocks using High Pure PCR Template Preparation Kit (Roche Life Science, Germany). The purification of desired DNA was calculated by determining the optical absorption ratio of 260 OD/280 OD and its concentration using the formula, μg/ml=50×P×OD260 (DNA dilution grade=P), by Biophotometer; moreover, to ensure the health of DNA, polymerase chain reaction with a housekeeping gene such as β-globin was performed. DNAs that did not pass the quality control stage and did not have a light absorption ratio of 1.6 to 2 and also a minimum concentration of 50 ml/μg were excluded from the evaluation.
Investigation of KRAS and BRAF gene mutations by reverse dot blot: To perform reverse dot blot (using RDB 2250 KRAS/BRAF Kit, AID Autoimmun Diagnostika GmbH, Germany), first, the multiplex polymerase chain reaction was performed on genomic DNA. The reaction mixture consisted of 15 µl PN-mix, 2.5 µl 10× PCR reaction buffer, 1.5 µl MgCl2, one unit of the single polymerase enzyme, and 5 µl DNA sample with a total volume of 25 µl.
The temperature program included an initial denaturation at 95°C for 5 min (One cycle). The polymerase reaction in the first step was performed at 95°C for 20 s, 60°C for 20 s and for 10 cycles. In the second step, it was performed at 95°C for 20 s, 55°C for 20 s, and 72°C for 20 s (26 cycles). Then, the final extension reaction was performed at 72°C for 8 min (One cycle).
For hybridization, first 20 μl of denaturation solution was mixed with 20 μl of PCR mixture, followed by incubation at room temperature for 5 min. The nitrocellulose strip was incubated in 1 ml of hybridization solution for 30 min at 47°C. The hybridization buffer was completely removed from the strip and the strip was washed twice with stringency wash solution, each time for 1 min. The nitrocellulose strip was then incubated in 1 ml of strignecy wash solution for 15 min at 47°C. The strip was washed twice at room temperature for 1 min by adding 1 ml of diluted rinse solution. Then, 1 ml of conjugate solution (Concentration of this solution was diluted 1:100 by conjugate buffer) was added to the strip, followed by incubation in a horizontal shaker at room temperature for 30 min. After removing the conjugate solution from the well, the strip was washed three times with 1 ml of rinse solution for 1 min. Afterward, 1 ml of the substrate solution, which reached room temperature, was added to the strip, followed by incubation for 10 to 20 min. After this period, the reaction was stopped by adding 1 ml of distilled water. After drying the strip, the results were interpreted using a nitrocellulose membrane scan.
Evaluation of KRAS gene mutations by PCR-RFLP method: Specific primers were designed and synthesized to identify codons 12 and 13 of the KRAS proto-oncogene using the NCBI bank and Blast software (RAS A: 5 ́ -ACTGAATATAAACTTGTGGTCCATG GAGCT-3 ́, RAS B: 5 ́ -TTATCTGTATCAAAGAAT GGTCCTGCACCA-3 ́,and RAS C: 5 ́-GGATGGTCC TCCACCAGTAATATGGATATT A-3 ́).
RAS A and RAS B primers were designed to synthesize 166 bp fragments in the first PCR and also RAS C primer along with RAS A primer was used to synthesize the second PCR product.
The first and second reaction mixtures consisted of 12.5 μl of amplicon master mix, 1 μl of each RASA (15 pmol) and RASB (15 pmol) primer for the first reaction mixture, 1 μl of each RASA (15 pmol) and RASC (15 pmol) primer for a mixture of the second reaction, and genomic DNA up to a concentration of 100 ng. The final volume of 25 μl was prepared.
The PCR reaction consisted of initial denaturation at 95°C for 4 min (One cycle), 30 cycles at 94°C for one min, 50°C for 1 min, and 72°C for 2 min. The final extension was performed in a cycle at 72°C for 5 min. To detect mutations in codons 12 and 13, the first PCR product was enzymatically digested with BSTX-I and XcmI enzymes (New England Biolabs, USA) for 24 hr according to the standard protocol. Also, to identify mutations in codons 12 and 13 of the second PCR product, enzymatic digestion was performed using BSTX-I and XcmI enzymes for 24 hr, respectively. The results of enzymatic digestion were evaluated on 12% polyacrylamide gel and by silver nitrate staining.
Evaluation of BRAF gene mutations by allele-specific PCR: In this method, two series of primers were used to detect the V600E mutation BRAF (F-wild type): 5’-TAGGTGATTTTGGTCTAGCTACCGT- 3’, BRAF (Reverse): 5’- GTAACTCAGCAGCATCTCAGGG- 3’, BRAF (F-mutant type): 5’- TAGGTGATTTTGGTC TAGCTACCGA- 3’). Accordingly, two separate mixtures were prepared for PCR.
The first and second reaction mixtures consisted of 12.5 µl of the master mix, one µl of each BRAF (F-wild type, 15 pmol) and BRAF (Reverse, 15 pmol) primer for the first reaction mixture and one µl of each ready-to-use primer BRAF (F-mutant type, 15 pmol and BRAF reverse, 15 pmol) for a mixture of the second reaction. Furthermore, genomic DNA was prepared up to a concentration of 100 ng and a final volume of 25 μl was prepared.
The PCR reaction consisted of initial denaturation at 95°C for 5 min (One cycle), 38 cycles at 95°C for 40 s, 58°C for 1 min, and 72°C for 20 s. The final extension was performed for one cycle at 72°C for 10 min. The reaction product was examined on 12% polyacrylamide gel and by silver nitrate staining.
Statistical analysis: The results were analyzed by SPSS software version 19 (IBM, USA). The chi-square test was applied to investigate the relationship between mutations and clinicopathological factors. The correlation test was used to investigate the relationship between mutations and factors with the normal distribution and p<0.05 was considered statistically significant.
Results :
Results of reverse dot blot: In the assessment of KRAS and BRAF gene mutations by reverse dot blot, each nitrocellulose strip had internal PCR control and conjugated buffer control for controlling hybridization; if there was no mutation in both oncogenes, only the band control could appear. If there was a mutation, the same area of the band could be observed depending on the type of point mutation (Figure 1). Due to transition and transgenic mutations, the wild-type sequence of GGT in codon 12 was mutated into c.35G> C (p.G12A), c.34G> C (p.G12R), c .35G> A (p.G12D), c.34G> T (p.G12C), c. [34G> A; 35G> T] (p.G12I), c. [34G> C; 35G> T] (p.G12I), c.34G> A (p.G12S), and c.35G> T (p.G12V).
The highest frequency of mutations was related to G12D (16%), and G12I and G12R mutations were not found in the study population. The frequency of mutations found in this study is shown in table 1.
Regarding mutations in codon 13 of the KRAS gene (Wild type GGC sequence), mutations of c.38G> A (p.G13D) and c.37G> T (p.G13C) were observed and G13C mutation was not found in this study; data shows the frequency of G13D mutation is equal to 12% (Table 1).
Results of PCR-RFLP: PCR-RFLP results about KRAS gene revealed the presence of mutations in the codons 12 and 13 of some samples of the studied patients. None of the enzymes could break the fragment if the fragment from the first PCR had a mutation. Fragments of 28 bp and 138 bp appeared by enzymatic digestion reaction in wild-type sequence. Moreover, if the amplified fragment from the second PCR had a mutation, three fragments of 134 bp, 106 bp, and 18 bp could be produced from the second PCR in mutable sequences, while bp18, bp106, and 28 bp fragments could be produced as a result of enzymatic digestion in normal type sequences (Figure 2). As shown in figures 2 and 3, 30% of samples showed a mutation in codon 12, and 12% of them had a mutation in codon 13 of KRAS oncogene.
Results of allele-specific PCR: To detect BRAF gene mutations, the presence of the allele of normal type and mutant type was investigated.
Figure 4 shows an example of polyacrylamide gel obtained from this study (Figure 4), in which the wild type and mutant alleles had a band at 180 bp position, and the interpretation of findings were done based on the results obtained from both types of alleles. Examining the frequency of V600E mutation in the BRAF gene revealed that out of 50 samples studied, 90% did not have V600E mutation, and only 10% showed V600E mutation.
Concordance of mutations analysis: The frequency of mutations found in codons 12 and 13 by the reverse dot blot method was consistent with the PCR-RFLP results, and also the results of V600E mutation in BRAF by the allele-specific PCR and reverse dot blot method were similar. A comparison of results among different methods is described in table 2. In this table, the differences between the methods in the evaluation of mutations are indicated. In reverse dot blot method, 10 point mutations in KRAS gene were examined by a mutation in the BRAF gene, and in addition to codons 12 and 13, codon 61 mutations in the KRAS gene were also examined. However, in RFLP method, only mutations in codons 12 and 13 were evaluated and mutation evaluation in BRAF gene was confirmed by allele-specific method.
Results of statistical analysis: Out of 50 samples, five had metastatic liver tissue of CRC origin, and other samples of cecum, colon, and rectum tissues had CRC (Table 1); out of 50 samples, 29 (58%) tumors metastasized, and in 21 (48%) samples, metastasis did not occur.
The relationship between sex and each of the mutations in KRAS and BRAF genes was determined using the Pearson's chi square test. There was no significant relationship between sex and type of mutations in these two genes.
Regarding this analysis, no significant relationship was found among the mutations in KRAS and BRAF genes and metastasis at the time of diagnosis (p>0.05). No significant relationship was found among the mutations in codon 12 of KRAS gene with high and low differentiation (p>0.05), and chi-square test showed no significant relationship among the mutations in KRAS 12 gene with mucinous and non-mucinous tumors. The results showed no significant relationship among the mutations in 13 KRAS gene with mucinous and non-mucinous tumors.
There was no significant association among the mutations in BRAF gene with mucinous and non-mucinous tumors.
Discussion :
KRAS oncogene is an important member of EGFR signaling cascade family, and KRAS activating mutations are among the most common mutations in human cancers 16. Due to the high prevalence of colorectal cancer in the world and the role of KRAS codons 12 and 13 in malignancy and tumor progression, timely screening is essential to identify patients at the early stages. Determining the type of mutation using non-invasive and rapid methods is also of considerable importance for targeted therapies appropriate to the type of mutation 17. The mutations in the codons 12 and 13 lead to changes in RAS protein which is resistant to GTPase activity; also, they result in long-term persistence of active form bound to GTP, thus leading to intensified cell growth and proliferation. BRAF and KRAS mutations both occur at the early stages of colorectal cancer but are rarely seen together 18. In a study by Velho et al in Portugal, Li et al in China, and Brink et al on Dutch patients, 35.3%, 37%, and 33.3% of patients had mutations in the KRAS oncogene, respectively 19-21. The most common mutations in KRAS gene are in codon 12 and include the amino acid substitutions, G13D, G12S , and G12D, of which G12D mutations are the most common 22. These results are consistent with results of our study because the highest frequency of amino acid substitution was found to be related to G12D (16%). In a study by Garcia-Carbonero et al on KRAS and BRAF mutations as predictors and predictive biomarkers in metastatic colon cancers, COX regression analysis showed that KRAS and BRAF in metastatic tumors are important agents for survival. Garcia-Carbonero et al stated that patients with KRAS mutations had a poorer response to chemotherapy 23.
In thyroid cancer, BRAF mutations were found to upregulate microenvironment genes, potentially increasing tumor invasion. The frequency of BRAF mutations is high in melanoma (44%) and lower in thyroid, colorectal, and ovarian cancers (8, 6, 5%, respectively) 24. In our study, the frequency of KRAS and BRAF oncogenes mutations in a population of CRC patients with a mean age of 56.96 was found to be 42% (30% in codon 12 and 12% in codon 13) and 10%, respectively. The highest number of samples with FFPE blocks belonged to male patients (68%, with a mean age of 56 years), while the lowest number of samples belonged to female patients (34%, with a mean age of 54.8 years) which is consistent to the study of Li et al. Among these patients, 15 had mutations in codon 12 of KRAS gene (30%), while 6 patients had mutations in codon 13 (12%).
Due to the results of this research and other studies, the highest frequency of mutations is related to codons 12 and then 13, although several studies indicated the role of codons 61 and 146 and their mutations such as codons 12 and 13; however, the frequency of mutations in codon 61 and 146 is lower than codons 12 and 13. Therefore, codons 61 and 146 also have a prognostic role in the treatment of patients with CRC, but the frequency of these mutations is very low compared to codons 12 and 13 which is consistent with our study findings showing the high frequency of mutations in codon 12 and also the low frequency of mutations in codon 61 25. The frequency of KRAS gene mutations in CRC patients is approximately 30 to 46% based on previous articles.
This wide range of frequencies reported from KRAS mutations is possible due to various factors, including the sensitivity and specificity of mutation detection methods, the small number of patients, or the variability in the region in which the gene was selected, namely codons 12, 13 and/or 61 regions 26.
Regarding the role of these two genes in improving CRC patients' outcomes, extensive studies have been performed to quickly and accurately evaluate these two genes in recent decades.
Techniques used for the mutant screening such as temperature gradient gel electrophoresis or SSCP and methods for mutant detection such as PCR-RFLP or allele-specific PCR amplification (MASA) may show differences in the sensitivity and/or mutation detection characteristics of the KRAS gene 27. Though the mutations were studied in a small population of patients, the selection of these patients was random and results were investigated by comparing three methods of reverse dot blot, PCR-RFLP, and allele-specific PCR.
The hybridization method was used to detect the mutations of codons 12, 13, 61 in the KRAS oncogene and also V600E mutation in the BRAF oncogene. Specific primers and probes were designed for each point mutation to detect them. Moreover, each nitrocellulose strip had internal controls for PCR accuracy and hybridization steps. However, all samples were re-analyzed by RFLP for the KRAS gene and allele-specific PCR for the BRAF gene. The results of these two methods confirmed each other, and the samples with mutations in each method were well distinguishable from the samples without mutations.
In this study, the mutations including G12A, G12R, G12D, G12C, G12S, and G12V in codon 12 and G13D, G13C in codon 13, Q61H in codon 61 of KRAS proto-oncogene, and V600E mutation in BRAF proto-oncogene were analyzed by reverse dot blot method. The mutations such as c.183A> C (p.Q61H), c.183A> T (p.Q61H), c.37G> T (p.G13C), and c.34G> C (p.G12R) were not found, and the highest frequency was related to G12D mutation (16%) of codon 12, which was studied and evaluated by Ince et al in 2005 28, Cabart et al in 2016 29, and Ergun et al in 2019 30. In the reverse dot blot method, point mutations in KRAS oncogene and also V600E mutation in BRAF oncogene were evaluated, and in the PCR-RFLP method, only the presence of mutations in codons 12 and 13 of KRAS was examined. The allele-specific PCR method was used to detect BRAF mutation in CRC. Hatzaki et al in 2001 presented a modified mutagenic PCR-RFLP method for the KRAS codon exon 1 mutations in tumor samples from NSCLC patients 31.
Some studies have reported the importance of detecting KRAS point mutations in CRC patients. De Roock et al and Tejpar et al reported that some specific mutations in KRAS can affect patients’ outcomes and patients with a mutation in codon 13 (G13D) have a survival benefit 32,33. Studies of Al-Mulla et al, Guerrero et al, and Smith et al on KRAS codon 12 mutations shows that patients with mutations in codon 12 compared to codon 13 may demonstrate carcinogenic potential as a result of apoptosis inhibition, loss of contact inhibition, and also increased contact-independent growth 34-36.
KRAS mutations have a prognostic role in CRC patients; in general, an association between KRAS codon 12 mutation (Specific G12V, G12C mutations) and overall survival was illustrated in patients with recurrent and metastatic colorectal cancer and wild type KRAS shows an increased overall survival 37.
KRAS G12D mutation shows a poorer prognostic effect on patients compared to the other mutations 38, and KRAS G12V shows a biological behavior in CRC patients 39.
Kaelin reported the synthetic lethality of KRAS, in which the mutations in two or more genes cause cell death 40 which is considered an advantage that leads to increased cytotoxicity against cancer cells without the toxicity associated with current chemotherapy 41.
Although the frequency of KRAS mutations in the hybridization report did not differ from PCR-RFLP results, point mutations in each codon were excluded in this study. Detecting point mutations is important in overall survival of patients. Besides, another method is required to examine BRAF mutation as the process of mutation analysis in KRAS and BRAF is very time-consuming.
As a result, the reverse dot blot method is a more sensitive and specific method to evaluate mutations in these oncogenes and PCR-RFLP and allele-specific PCR methods seem to be suitable for screening the existence of mutation in KRAS and BRAF oncogenes.
Allele-specific PCR is an optimal method to evaluate V600E mutation in other cancers in which BRAF alone is a biomarker for detecting melanoma, thyroid, and ovarian cancer. In the studies of Osumi et al 42 and Chen et al 43, the effect of mutation type on anti-EGFR therapies was investigated; therefore, point mutation analysis plays an essential role in treating patients with CRC.
Conclusion :
Due to high frequency of mutations, the young age at diagnosis, and the oncogenes’ roles in disease management, it is recommended to study the mutations of KRAS and BRAF oncogenes in patients with colorectal cancer. The results of PCR-RFLP and allele-specific PCR were similar to reverse dot blot, although PCR-RFLP and allele-specific PCR are time-consuming methods. These findings suggest that PCR-RFLP and allele-specific PCR as confirmatory methods are suitable for screening the existence and presence of the mutation in KRAS and BRAF oncogenes. Furthermore, for a more accurate assessment and also discovering the possible role of the type of mutations, another method with more sensitivity is needed. In general, the reverse dot blot method was faster and more sensitive to detect KRAS and BRAF mutations; moreover, type of KRAS mutations could be assessed which saves response time, reduces additional costs, and provides higher quality services. Therefore, in the current study, the superiority of the reverse method over the RFLP method was proved.
Acknowledgement :
This article was extracted from a master thesis entitled "Evaluation of KRAS & BRAF mutations in patients suffering from colorectal cancers" in cellular and molecular biology field and approved by Mazandaran University and performed in Payvand clinical laboratory.
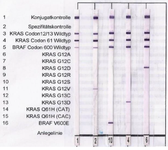
Figure 1. Nitrocellulose strips from reverse dot blotting. Results from right: G12D mutation, G13D mutation, V600E mutation, G12V mutation, no mutation in both oncogenes.
|

Figure 2. Polyacrylamide gel. Codon 12 KRAS has two bands at the 134 bp and 106 bp sites, indicating a mutation. Bands at 106 bp had no mutations in this codon: Lane 1: a 100 bp marker, Lane 3 and Lane 5: no mutations, and other lanes: mutations.
|

Figure 3. Polyacrylamide gel of the 13 oncogene KRAS codon. Dual-band samples at bp134 and bp106 sites have mutations in this codon, and bands at 106 bp site have no mutations in this codon: Lane 9: 20 bp marker, Lane 8 100 bp marker, Lanes 5, 6, and 7: no mutations and Lanes 1, 3, and 4: mutations.
|
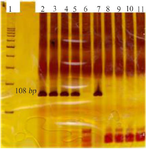
Figure 4. BRAF gene polyacrylamide gel. Lane1: 100 bp marker, Lane 2 to Lane 6: mutant alleles, and Lane 7 to Lane 11: non-mutant alleles. It is interpreted as meaning that the first sample has a mutation and the other three samples do not have a mutation. Lane 6 and 11 are PCR (M.M) mixture contamination controls.
|
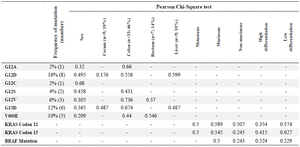
Table 1. Correlation of mutation status with clinical and histopathological features
|
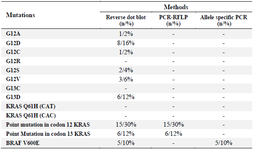
Table 2. Comparison of results among different methods
|
|