Association of a Functional Single Nucleotide Polymorphism (rs874040) in the RBPJ Gene with Susceptibility to Rheumatoid Arthritis in Iranian Population
-
Salesi, Mansour
-
Department of Internal Medicine, Faculty of Medicine, Isfahan University of Medical Science, Isfahan, Iran
-
Oboodiyat, Mahdieh
-
Department of Internal Medicine, Faculty of Medicine, Isfahan University of Medical Science, Isfahan, Iran
-
Salehi, Rasoul
-
Department of Genetics and Molecular Biology, Faculty of Medicine, Isfahan University of Medical Sciences, Isfahan, Iran
-
 Pakzad, Bahram
Department of Internal Medicine, Faculty of Medicine, Isfahan University of Medical Science, Isfahan, Iran, Tel:+989133120418; Email: bpakzadd@yahoo.com
Pakzad, Bahram
Department of Internal Medicine, Faculty of Medicine, Isfahan University of Medical Science, Isfahan, Iran, Tel:+989133120418; Email: bpakzadd@yahoo.com
Abstract: Background: Rheumatoid Arthritis (RA) is a progressive, heterogeneous, and common multifactorial autoimmune disease. Several Genome-Wide Association Studies (GWASs) have revealed more than 100 risk loci for RA. One of these loci is a functional single nucleotide polymorphism (rs874040; G>C) near the recombination signal-binding protein for the immunoglobulin kappa J region (RBPJ) gene. RBPJ can convert into a transcriptional activator upon activation of the canonical Notch pathway. Notch signaling has recently emerged as an important regulator of immune responses in inflammation and autoimmune diseases. In the present study, the possible association between SNP rs874040 (G>C) upstream of the RBPJ gene with RA risk was assessed in Iranian population.
Methods: A case-control study including 60 RA patients and 44 control subjects was conducted to estimate rs874040 genotypes using real‑time polymerase chain reaction High Resolution Melting (HRM) method.
Results: Logistic regression analysis indicated that homozygous CC and heterozygous GC genotypes increase the risk of RA compared with GG genotype (CC vs. GG; OR=11.36; 95% CI [3.93-33.33] and CG vs. GG; OR=3.78; 95% CI [1.30-10. 98]). Besides, subjects with C allele were more frequently affected with RA than subjects with G allele (OR=10.42; 95% CI [5.21-20.83]). Furthermore, in the patient group, a significant correlation was found between C-reactive protein concentrations and rs874040 polymorphism (p<0.05).
Conclusion: Our findings propose a substantial correlation between rs874040 polymorphism and RA risk in Iranian population.
Introduction :
Rheumatoid Arthritis (RA) is a progressive, heterogeneous, common, complex, multifactorial autoimmune disease 1,2. It causes inflammation along with painful swelling in the affected parts of the body, especially in joints. RA commonly affects the small joints such as hands, knees or ankles, and typically the same joint on both sides of the body; this can eventually result in stiffness, bone and cartilage erosion, and joint deformity 3,4. The global annual incidence of RA is around 3 cases per 10,000 population and its prevalence is about 1% 5-7. RA is a multifactorial disorder with the involvement of multiple genetic and environmental factors 8,9. Several researchers have propounded a significant genetic susceptibility for RA with 50-60% heritability, suggesting that a large proportion of the RA could be the result of underlying genetic risk factors 10. This notion was strengthened with high concordance rates in monozygotic twins (12.3-15.4%) by analogy with dizygotic twins (3.5%) 11. During the last two decades with advances in genotyping methods such as Genome-Wide Association Studies (GWAS), more than 100 genetic susceptibility loci associated with RA risk have been obtained 12,13. One of these loci is a single nucleotide polymorphism (rs874040; G>C) near recombination signal-binding protein for immunoglobulin kappa J region (RBPJ) gene. Considering the previous studies, RBPJ is a transcriptional repressor but converts into a transcriptional activator upon activation of the canonical Notch pathway 14. Numerous studies found that Notch signaling regulates multiple steps of T and B cell development in the immune system. Notch pathway regulates differentiation of lineages of lymphoid T and B cells, T helper cell differentiation as well as T cell activation, and regulatory T cell function 15-17. This pathway partakes in the expression regulation of inflammatory cytokines that are involved in RA pathogenesis 18,19. Furthermore, in normal situations, by the effect of some cytokines such as transforming growth factor (TGF)-b1 and IL-6, naive CD4+ T cells are differentiated into Th17 cells and express IL-10 and IL-17 and could not prompt autoimmunity 20. However, under the effect of IL-23 and IL-23R, Th17 cells could produce proinflammatory cytokines and induce autoimmunity 21.
In RA, RBPJ expression is increased in memory CD4+ T cells extracted from RA patients compared with the control group, and this elevated expression was related to prompted inflammatory cytokines such as IL-17A, IL-9, and IFNγ in response to activation of Notch signaling 22. Previous studies reported that CD4+ T cells and especially memory CD4+ pooled into the RA synovium are involved in synovial inflammation by increasing the production of proinflammatory cytokines such as TNF-α, IL-1β, IL-6, and IL-17 from macrophages, synovial fibroblasts, and other available leukocytes 23. Orent et al demonstrated that rs874040 is a functional SNP that is located in a strong enhancer region containing DNase I hypersensitive sites for RBPJ. CC genotype in this SNP increased the expression of the RBPJ gene in stimulated memory CD4+ T cells in normal subjects compared with persons with GG as a protective genotype 22. Hence, it seems that this functional variant has a critical role in increasing the susceptibility to RA disease. In the present study, the possible association between SNP rs874040 (G>C) upstream of the RBPJ gene with RA risk was evaluated in Iranian population for the first time.
Materials and Methods :
Study population and sample preparation: This case-control study comprised a total of 104 individuals that were selected from subjects referred to Alzahra Hospital which is the largest affiliated hospital of Isfahan University of Medical Sciences. Subjects included 60 unrelated persons with RA disease (including 35 females and 25 males) who met the diagnostic criteria created by the American College of Rheumatology (ACR) and 44 unrelated healthy subjects (including 21 females and 23 males) who had no symptoms or personal and family history of RA or other autoimmune disorders.
All participants signed an informed consent form and the Ethics Committee of the University approved the study. The participants were interviewed and demographic data on sex, age (during sampling period) and age of onset, Body Mass Index (BMI, calculated as weight [kg] divided by height [m] squared), blood pressure, the presence of Diabetes Mellitus (DM), thyroid disease and family history of RA and other autoimmune conditions were obtained using a structured questionnaire. Also, laboratory indices including Erythrocyte Sedimentation Rate (ESR), C-reactive protein (CRP), White Blood Cell (WBC), hemoglobin, Platelet count test (PLT), creatinine, Blood Urea Nitrogen (BUN), Fasting Blood Sugar (FBS), High-Density Lipoprotein (HDL), Low-Density Lipoprotein (LDL), and Triglyceride (TG) were recorded.
Genotyping of polymorphism: Genomic DNA was extracted from 2 ml ethylenediaminetetraacetic acid (EDTA)-anticoagulated peripheral blood samples by PrimePrep Genomic DNA Isolation Kit (GeNetBio, Korea). The quality and concentration of all genomic DNA samples were assessed by agarose gel electrophoresis and spectroscopy, respectively, and then DNA was stored at −20°C until genotyping by real‑time polymerase chain reaction High‑Resolution Melting (HRM) method. HRM was performed using HOT FIREPol EvaGreen HRM Mix (no ROX) HRM PCR kit that contains HOT FIREPol® DNA Polymerase, 5x EvaGreen® HRM buffer, 12.5 mM MgCl2, dNTPs, bovine serum albumin (BSA), and EvaGreen dye (Solis BioDyne, Estonia). Analysis was carried out by HRM on a Rotor-Gene 6000 (Corbett Research, Australia). In this method, polymorphism in the PCR product is detected by changes in the shape of the melting curve compared to a reference sample. Amplification of fragment (141 bp) was performed using the primer sense (CGCTACAGTGGTGACCCC) and antisense (GTCATCTCCACCTGCCCATA). A 35-cycle PCR was performed with the following conditions: 5 min at 95°C for denaturation of the template DNA for the first cycle, denaturation at 95°C for 20 s, annealing at 60°C for 30 s, and extension at 72°C for 20 s. The melting curve is generated by increasing the temperature between 65°C and 95°C at 0.1°C/s heating rate. The melting curve is produced by the reduction in fluorescence with the increase in the temperature; nucleotide changes result in different curve patterns. For using sample genotypes in HRM analysis as a standard, specific samples (with different curves) were subjected to direct Sanger sequencing and their correct genotypes were determined.
Statistical analyses: SPSS V22 (IBM Corp, NY) was used for statistical analyses. The allele and genotype frequencies of rs874040 were tested for Hardy Weinberg equilibrium using chi-squared test. Logistic regression analysis was conducted to investigate the association between genotypes and RA and calculate specific Odds Ratios (ORs), 95% Confidence Intervals (CIs), and p-values. For demographic, clinical, and laboratory characteristics, p-values were calculated using independent sample t-test, Chi‑square, or Mann–Whitney test. The significance level was set at p<0.05.
Results :
In our analysis, a total of 60 patients (mean age: 50.30±10.55) and 44 controls (mean age: 48.73±10.93) were evaluated for RBPJ (rs874040) polymorphisms. Table 1 shows the characteristics of RA patients and healthy controls. In the RA patient group, the mean age of onset was 41.58±10.85. There was no important association between case and control groups regarding age (p=0.46) and sex (p=0.55), illustrating that for these factors matching was acceptable. Based on our results, there was a statistically significant difference in terms of Body mass index (BMI) (p<0.001) and positive family history of RA and other autoimmune diseases (p=0.002). As demonstrated, just 7 (11.7%) patients had DM whereas controls did not have DM (p=0.02). On the other hand, there was no significant difference between the patients and healthy subjects in the blood pressure parameter (p>0.05). Results of laboratory tests revealed that the concentration of some factors such as ESR, CRP, PLT, creatinine, and BUN was significantly higher in patients than in healthy controls (p<0.05). But the concentration of hemoglobin was significantly lower in patients than in healthy controls (p<0.001). The details of the laboratory characteristics of patients with RA and control groups are presented in table 2.
Genotype and allele distribution: Our findings demonstrated that the genotype distribution of rs874040 polymorphism in case and control groups was in agreement with Hardy Weinberg equilibrium. Among the RA cases, the frequency of GG, GC, and CC was 15, 28.3 and 56.7%, respectively. In the control group, the frequency of rs874040 genotype was 54.5% for GG, 27.3% for GC, and 18.2% for CC. Our study elucidated the significant association between CC (Compared with GG; p<0.001) genotypes and RA risk. Also, there was a significant difference between the heterozygote genotype (GC) compared with the GG genotype in the increased risk of RA (p=0.012).
Comparing the combined genotype, our results unveiled that the CC+GC (85% in cases and 15% in control groups) compared to the GG genotype (45.5% in cases and 54.5% in control groups) increased the risk of RA (p<0.001).
Likewise, concerning allele distribution, it was found that the C allele had a high frequency in the case group (80.95%) compared to the G allele (33.33%) and our assessment showed that the C allele was correlated with increased risk of RA (p<0.001). Moreover, our investigation revealed that stratification based on the median concentration of CRP in the patient group is significantly different in genotype groups (p<0.05). In fact, patients with risk allele (C) had a higher concentration of CRP. However, there was no significant association between the stratification by the age of onset, gender, BMI, ESR, and creatinine concentrations with this polymorphism (p>0.05) (Table 3).
Discussion :
It goes without saying that a large number of Single Nucleotide Polymorphism (SNP) loci affect susceptibility to RA. Genome-wide association studies reported more than 100 genetic susceptibility loci associated with increased risk of RA 12,13. Stahl et al carried out a GWAS on 5,539 subjects with RA and 20,169 controls of European descent and reported that rs874040 (CC genotype) upstream of the RBPJ gene was associated with an increased risk of RA 24.
On the other hand, Notch signaling has recently emerged as an important regulator of immune responses in inflammation and autoimmune diseases because this signaling pathway takes part in the regulation of multiple steps of T and B cell development 15,25,26. After activation of Notch receptors, Notch Intracellular Domain (NICD) is translocated to the nucleus where it interacts with RBPJ protein via the RAM domain and activates the expression of inflammatory cytokines. Meanwhile, RBPJ without activation of the Notch receptor is bound to specific DNA binding sites and is thought to act as a transcriptional repressor 15,27.
Horste et al demonstrated that RBPJ directly increases the expression of IL-23R by binding to the Il23r promoter and eventually represses anti-inflam-matory IL-10 production in the Th17 cells 28. Orent et al initially reported that in patients with RA, expression of RBPJ is increased in memory T cells compared with control subjects. Then, they revealed that the CC genotype of the rs874040 variant in memory CD4+ T cells leads to increased expression of the RBPJ gene compared with the GG genotype 22.
To the best of our knowledge, this study is the first research in the Iranian population that investigated the association between RBPJ polymorphism named rs874040 with the increased risk of RA. In our study, logistic regression analysis showed that CC genotype increases the risk of RA in comparison with the GG (CC vs. GG; OR=11.36; 95% CI [3.93-33.33]). In addition, heterozygote genotype (GC) increased the risk of RA by analogy to GG genotype (GC vs. GG; OR=3.78; 95% CI [1.30-10. 98]). Furthermore, subjects with the C allele were more frequently affected with RA than those with the A allele (OR=10.42; 95% CI [5.21-20.83]) (Table 4). Our results were consistent with two GWAS results conducted in two different populations; Stahl et al worked on European populations and Aslam et al investigated possible associations with RA risk in Pakistani population 24,29.
Moreover, in our patient group, a significant correlation between CRP concentration and rs874040 polymorphism (p<0.05) was found (Table 3). The amount of this factor indicates levels of inflammation in the body and alludes to active disease. This result demonstrates the association of risk allele with the severity of the disease (normal amount of CRP was less than 10 mg/L).
Conclusion :
This investigation disclosed a significant association between rs874040 polymorphism with RA risk in the Iranian population; however, probably some possible limitations exist in the study as the threats to the validity of our results such as small sample size. It seems that further association studies with a larger sample size would help to confirm the suggested correlations. Besides, other variants that were not included in our study might be involved in determining the risk of RA.
Acknowledgement :
We would like to appreciate any support provided by Isfahan University of Medical Sciences.
Conflict of Interest :
There is no conflict of interest to declare.
Conflict of Interest :
There is no conflict of interest to declare.
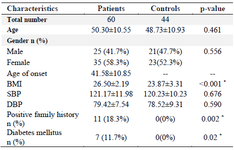
Table 1. Baseline characteristics of RA patients and control subjects
Data are represented as mean±SD, or n (%). * p<0.05. RA: Rheumatoid Arthritis; BMI: Body Mass Index; SBP: Systolic Blood Pressure; DBP: Diastolic Blood Pressure.
|
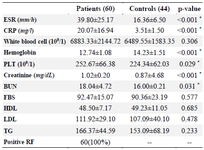
Table 2. Laboratory characteristics of patients with RA and controls group
Data are represented as mean±SD, or n (%).* p<0.05. RA: Rheumatoid Arthritis; ESR: Erythrocyte Sedimentation Rate; CRP: C‑Reactive Protein; BUN: Blood Urea Nitrogen; PLT: Platelet; HDL: High‑Density Lipoprotein; LDL: Low‑Density Lipoprotein; TG: Triglyceride; FBS: Fasting Blood Sugar; SD: Standard Deviation.
|
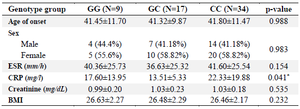
Table 3. Stratification analyses of the RBPJ polymorphism (rs874040) in patients
Data are represented as mean±SD, or n (%). * p<0.05. ESR: Erythrocyte Sedimentation Rate; CRP: C‑Reactive Protein; BMI: Body Mass Index; SD: Standard Deviation.
|
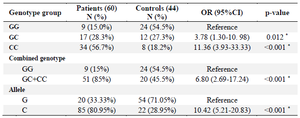
Table 4. Association between genotypes and allele frequency with RA risk
* p<0.05.
|
|