The Novel Drug Discovery to Combat COVID-19 by Repressing Important Virus Proteins Involved in Pathogenesis Using Medicinal Herbal Compounds
-
Mahmoudi, Samira
-
Department of Microbial Biotechnology, Faculty of Biological Sciences, Tehran North Branch, Islamic Azad University, Tehran, Iran
-
Balmeh, Negar
-
Department of Cell and Molecular Biology, Faculty of Biology, Nour Danesh Institution of Higher Education, Meymeh, Iran
-
Mohammadi, Niloofar
-
Department of Sciences and Agricultural Engineering, School of Agricultural Sciences, Pir Bakran Branch, Payame Noor University,, Isfahan, Iran
-
 Sadeghian-Rizi, Tahereh
Department of Biotechnology, Faculty of Biological Sciences and Technology, Shahid Ashrafi Esfahani University, Isfahan, Iran , Tel: +98 9132366325; E-mail: t.sadeghian@ashrafi.ac.ir
Sadeghian-Rizi, Tahereh
Department of Biotechnology, Faculty of Biological Sciences and Technology, Shahid Ashrafi Esfahani University, Isfahan, Iran , Tel: +98 9132366325; E-mail: t.sadeghian@ashrafi.ac.ir
Abstract: Background: The cause of COVID-19 global pandemic is SARS-CoV-2. Given the outbreak of this disease, it is so important to find a treatment. One strategy to cope with COVID-19 is to use the active ingredients of medicinal plants. In this study, the effect of active substances was surveyed in inhibiting four important druggable targets, including S protein, 3CLpro, RdRp, and N protein. RdRp controls the replication of SARS-CoV-2 and is crucial for its life cycle. 3CLpro is the main protease of the virus and could be another therapeutic target. Moreover, N protein and S protein are responsible for SARS-CoV-2 assembly and attaching, respectively.
Methods: The 3D structures of herbal active ingredients were prepared and docked with the mentioned SARS-CoV-2 proteins to obtain their affinity. Then, available antiviral drugs introduced in other investigations were docked using similar tools and compared with the results of this study. Finally, other properties of natural compounds were uncovered for drug designing.
Results: The outcomes of the study revealed that Linarin, Amentoflavone, (-)-Catechin Gallate and Hypericin from Chrysanthemum morifolium, Hypericum perforatum, Humulus lupulus, and Hibiscus sabdariffa had the highest affinity for these basic proteins and in some cases, their affinity was much higher than antiviral medicines.
Conclusion: In addition to having high affinity, these herb active ingredients have antioxidant, vasoprotective, anticarcinogenic, and antiviral properties. Therefore, they can be used as extremely safe therapeutic compounds in drug design studies to control COVID-19.
Introduction :
Coronavirus disease 2019 (COVID-19) emerged in late 2019 by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and posed a global public health emergency. SARS-CoV-2 is a new coronavirus and the World Health Organization (WHO) firstly named it 2019-nCoV 1,2. The reason for naming coronavirus SARS-CoV-2 is the similarities of its genomic sequence to severe acute respiratory syndrome coronavirus (SARS-CoV) and the disease was subsequently called COVID-19 3. Coronaviruses, a genus of the Coronaviridae family, are classified as single-stranded, positive-sense RNA viruses, and their genomes range from 25-32 kb 4. According to the results of genome-based phylogenetic analysis, SARS-CoV-2 is a new branch of the betacoronavirus genus. SARS-CoV and the Middle East Respiratory Syndrome (MERS) are also from the betacoronavirus genus. It is noteworthy that SARS and MERS also appeared in 2002 and 2012, respectively 5,6. SARS-CoV caused an epidemic in China and MERS began in Saudi Arabia 7. The genome of SARS-CoV-2 codes for Nonstructural Proteins (NSPs) such as 3-chymotrypsin-like protease (3CLpro), RNA-dependent RNA polymerase (RdRp), accessory proteins, and Structural Proteins (SPs) 8. SPs include Nucleocapsid (N) protein, Membrane (M) protein, Envelope (E) protein, and Spike (S) protein 9.
The S protein is responsible for virus-cell interactions during viral entry 10. S1 and S2 are the two subunits of the S protein. S1 is at the N- terminus and responsible for the virus–receptor binding while S2 is at the C-terminus and responsible for virus-cell membrane fusion 11,12. S1 itself is divided into two domains called the Receptor-Binding Domain (RBD) and the N-Terminal Domain (NTD) 7. In fact, SARS-CoV-2 attaches to cells by its S protein and human Angiotensin-Converting Enzyme 2 (ACE2) 13-15. This binding triggers a cascade of events and leads to a fusion between viral and cellular membranes. Then, RNA viral genome is released into the cytoplasm of host cells 16. The next step is synthesizing polyproteins for encoding the viral replicase-transcriptase complex 17. So, targeting S protein could be an initial solution to prevent the infection.
The N protein is in the helical nucleocapsid and has an important role in replication, RNA packaging, and pathogenesis 4. N protein is more conserved compared to other SPs and more expressed throughout COVID-19 infection; thus, it is one of the immunodominant antigens in SARS-CoV-2 18,19. Therefore, N protein inhibition seems to be one of the fastest and most effective actions to prevent the infection progression in patients infected with COVID-19.
During transcription of the SARS-CoV-2 genome, the virus produces a ~800 kDa polypeptide and cleaves this polypeptide by 3CLpro to generate NSPs 20. 3CLpro converts this polypeptide to eleven fragments. The 3CLpro is located at the 3' end of the genome that has excessive variability 21. In fact, 3CLpro has a critical role in the virus particles' replication so this protein is a potential target for inhibiting SARS-CoV-2 22.
RdRp is another important protein for SARS‐CoV‐2 with a range of 240-450 kDa and a catalytic core with differentiated domains 23. RdRp catalyzes the RNA replication from its RNA templates, i.e., the viral RNA is synthesized by RdRp. The sequences of RdRp in SARS-CoV, SARS-CoV-2, and MERS-CoV are almost similarly encoded 24. RdRp active site is a deep and huge groove for polymerization of RNA 25. This active site is the most accessible and conserved region, so targeting this region may be an effective therapeutic target to inhibit viral replication 26.
SPs and NSPs are both promising targets to inhibit the virus; while SPs are important in the infection processes, NSPs are involved in virus replication and transcription 27.
Many of the available antiviral drugs undergoing clinical trials are actually being tested for repurposing, such as chloroquine and Hydroxychloroquine, which are used to cure arthritis and malaria, respectively 28, and are currently being used in patients with COVID-19 29. Multiple bioinformatics studies have even been performed to find the inhibitory effects of various antiviral chemicals on SARS-CoV-2. For instance, in an in silico study, a drug-repurposing investigation was performed on quinoline-based inhibitors against three proteins, S protein, 3CLpro, and RdRp. The results exhibited that Saquinavir and quinoline,1,2,3,4-tetrahydro-1-[(2-phenylcyclopropyl)sulfonyl]-trans-(8CI) inhibited 3 CLpro, oxolinic acid and elvitegravir inhibited RdRp, and rilapladib inhibited S protein 30. But no therapeutic drug has yet been approved by the FDA that is applied exclusively for these patients 29. As a result, it is necessary to perform structural based analyses to identify inhibitors for SARS-CoV-2 proteins.
In recent years, medicinal plants have attracted significant attention to treat infectious diseases. Complex molecular structures and a wide variety of natural compounds make medicinal plants an excellent biological resource for drug discovery. So far, a limited number of plant species have been used for medicinal purposes, so there is much hope for their increasing use in the future 31. Flavonoids are natural compounds that have a wide range of functions, including promoting general immunity, blocking transcription and translation of pathogenic microorganisms, and being non-toxic to human cells. These factors have led to the notion of flavonoids as antiviral agents in various articles and studies 32. A laboratory study examined the inhibitory effect of flavonoid compounds and found that pectolinarin, herbacetin, and rhoifolin strongly blocked the 3CLpro of SARS-CoV 33. Thus, in this article, the inhibitory effect of active ingredients of different medicinal plants was investigated on four important virus proteins including S protein, N protein, 3CLpro, and RdRp. These plant-based substances have characteristics such as fewer side effects, cost-effectiveness, and greater availability. The results of this in silico approach showed that some of these studied active ingredients have a high affinity for each of the four important viral proteins compared to the inhibitors previously reported for each of these proteins. They could possibly have an inhibitory effect on the SARS-CoV-2 virus and COVID-19.
Materials and Methods :
Modeling and structural analysis: After obtaining all information about SARS-CoV-2 structure and genome through the ViralZone database (www.viralzone.expasy.org) 34, the sequences of S protein, 3CLpro, RdRp, and N protein were acquired from the NCBI database (https://www.ncbi.nlm.nih.gov/) 35 and modeled by the SWISS-MODEL database (https://swissmodel.expasy.org/) 36. Also, the RCSB PDB database (https://www.rcsb.org/) was used to download the SARS-CoV-2 structure 37.
Collecting the active substances of medicinal herbs: In this study, the active ingredients of medicinal plants were obtained from articles that had previously introduced the active ingredients in each plant using the GC-MS technique. Plants were selected based on their potential or approved antimicrobial properties as well as the information about their active ingredients. Criteria for selecting effective ingredients included having an equal affinity or greater than the available inhibitors to the mentioned proteins (Table 1).
Then, 3D structures of medicinal herbal substances were obtained from the PubChem database (https:// pubchem.ncbi.nlm.nih.gov/) 38.
Molecular analysis: These obtained 3D structures and that of viral proteins were prepared by docking methods through removing optimal H-bond and all water and adding Gasteiger charges. Then, molecular docking analysis was conducted between each herbal compound and viral proteins through AutoDock vina. Figures were created by Chimera software, version 1.14 39. Also, to ensure the suitability of the proposed herbal compounds in this study, the blockers and antiviral drugs introduced in other studies were docked with selected viral proteins.
Investigation of characteristics of high scoring compounds; Finally, Way2Drug (www.pharmaexpert.ru/pass online/predict.php) 40, lazar (lazar.in-silico.ch/predict) 41, and AVCpred (http://crdd.osdd.net/servers/avcpred) 34 were used to obtain the activity, daily dose carcinogenicity, blood-brain barrier penetration, and antiviral features, respectively.
Results :
The best herbal compounds for S protein, 3CLpro, RdRp, and N protein were recognized based on the highest binding affinity. The affinity of reported viral proteins’ blockers was obtained and compared with the top scoring natural compounds examined in the present study. The results presented in table 2 have shown that top scoring medicinal herbal compounds for 3CLpro and RdRp had more affinity compared with previously reported blockers. Also, information on all plant chemicals docked with N, 3CL, S, and RdRp proteins is given in tables 1-4, respectively.
The most common medicinal herbal compounds for all mentioned viral proteins were investigated in this study. Among them, Amentoflavone had the highest binding energy with the scores of -8, -8.9, and -9.4 Kcal/mol for S protein, 3CLpro, and RdRp, respectively, and Linarin was the best top scoring compound for interaction with SARS-CoV-2 N protein (Score of -8.1 Kcal/mol) (Table 3).
The important amino acids involved in the binding of mentioned viral proteins that were achieved from previous articles and the interaction of H-bond related to top scoring compound for blocking each viral protein are shown in figure 1.
Based on the lazar database, characteristics of common compounds including respiratory analeptic, antitussive, antivirus, RNA synthesis inhibitor, and hepatoprotection were investigated. Other specifications such as carcinogenicity, blood-brain barrier penetration, herbal compounds antiviral ability, and maximum recommended daily dose were examined and indicated in table 4.
Discussion :
At present, researchers around the world are examining several different technologies, medicines, and vaccines against COVID-19. So far, ten candidate vaccines have been administered to volunteers in safety trials 42, among which one has reached the second phase of human trials 43. However, there is still no conclusive evidence that vaccines can make people completely safe; so along with vaccine production, special attention should be paid to therapeutic drug development 44.
Blocking the steps of the viral lifecycle provides potential targets to cure COVID-19 infection. SARS-CoV-2 has two types of proteins, including SPs and NSPs. Both proteins are important for virus survival and the infection process, so preventing the action of these proteins could be helpful to create anti-SARS-CoV-2 drugs 45. Inhibition of S and N proteins can be a quick solution for preventing SARS-COV-2 infection 46 and further, given that 3CLpro and RdRp are involved in virus replication and development, it is necessary to identify novel compounds that can repress the 3CLpro and RdRp 47,48.
There are two strategies for preventing SARS-CoV-2 interaction with the host cell. Blocking virus receptors (ACE2, TMPRSS2, and GRP78) on the surface of host cells and inhibiting virus proteins are the two methods. In our previous study, the effect of 1032 medicinal herbal compounds on inhibiting SARS-CoV-2 receptors was investigated and based on the results, Berbamine, Hesperidin, and Hypericin had the highest affinity to target receptors of S protein 49. Subsequently, in the present study, previous investigations were extended to screen various medicinal herbal compounds against S protein, 3CLpro, RdRp, and N protein of the virus.
Some studies stated that a few chemical drugs such as Colistin, Nelfinavir, and Prulifloxacin could be used to target SARS-CoV-2 3CLpro 50,51. One study examined the inhibitory effect of existing antiviral compounds. This research indicated the interaction of about 123 antiviral drugs against 3CLpro and Paritaprevir, Raltegravir, Bictegravir, and Dolutegravir 52. In an investigation, a medicinal plant database containing nearly 32,297 antiviral phytochemicals was screened, and outcomes approved that 5,7,30,40-tetrahydroxy-2’-(3,3-dimethylallyl) isoflavone had the highest affinity to 3CLpro 22. An in silico study performed computational drug design methods to determine inhibitors of 3CLpro from available FDA approved antiviral medicines. As a result, three antiviral drugs including Saquinavir, Remdesivir, Darunavir, and two natural compounds including coumarin derivatives and flavone were promising hits and can inhibit SARS-CoV-2 3CLpro function 53.
Based on a molecular dynamics study, inhibitors were tested against SARS-CoV-2 RdRp and it was revealed that Ribavirin, Sofosbuvir, Remdesivir, Galidesivir, Cefuroxime, Favipiravir, Hydroxychloroquine, and Tenofovir could tightly attach to the RdRp active site. Also, they presented that YAK, Setrobuvir, and IDX-184 can tightly wrap to RdRp and eradicate the virus 48. In addition to many studies performed to detect the inhibitory effect of chemical compounds on RdRp protein, research has been done to find the effect of traditional Chinese medicinal compounds on this protein. For instance, a research group screened some medicinal compounds and found that Theaflavin had lower binding energy while docking with SARS-CoV‐2 RdRp, thus, Theaflavin can be a potential inhibitor for it 25. Since the advent of SARS-CoV-2, much research has been done on the inhibitory effect of Remdesivir on RdRp 54 and ultimately, its effect was approved by FDA 55. A study detected top natural compounds by virtual screening so that Loniflavone and Simeprevir had the highest affinity to S protein; also, Amyrin and Conivaptan had the highest affinity to the N protein 56.
According to the results of the present study, Linarin with the score of -8.1 kCal/mol, Catechin Gallate with the score of -9.2 kCal/mol, Amentoflavone with the score of -8, and Amentoflavone with the score of -9.4 kCal/mol had a higher affinity for N protein, 3CLpro, S protein, and RdRp, respectively. Also, the active ingredient Amentoflavone obtained from Hypericum perforatum plant had a relatively high affinity for all four important proteins of SARS-CoC-2, so this active ingredient may prevent the binding, entry, multiplication, and development of the virus.
Comparison of the binding affinity of previously reported blockers for each four mentioned viral proteins and top scoring herbal compounds investigated in this survey revealed that the compounds obtained in this study have a greater tendency to bind to virus proteins in some cases. The antiviral drug Prulifloxacin had a score of -8.3 kCal/mol for binding to the 3CLpro protein, while (-)-Catechin Gallate had by far more tendency to bind to this protein, with a score of -9.2 kCal/mol. In the case of RdRp, the antiviral drug Theaflavin had the -9.3 kCal/mol docking score, while Amentoflavone had the -9.4 kCal/mol docking score. It should be noted that Remdesivir, which has been already approved by the FDA to inhibit RdRp protein, has a much lower score (-7.8 kCal/mol) than Amentoflavone.
In addition to having fewer side effects, Linarin and Amentoflavone are not carcinogenic. Linarin and Amentoflavone have other properties as they are antioxidants, vasoprotective, anticarcinogenic, and antiviral, which make them an extremely safe therapeutic compound in the respiratory tract.
According to our previous study on the effect of herbal active ingredients in inhibiting receptors of SARS-CoV-2 and their effect on viral proteins in the present study, it was found that Hypericin as an herbal compound has a high affinity for ACE2, TMPRSS2, and GRP78 receptors and four viral proteins, thus, it can be an important blocker/inhibitor for SARS-CoV-2 proteins and target cell receptors in treatment and control of COVID-19. Therefore, laboratory studies of this effective substance are a great help in discovering the proper medicine for the epidemic disease.
Conclusion :
Most studies have examined one or two proteins of SARS-CoV-2, but in this research, the inhibitory effect of medicinal herbal compounds was investigated on S protein, N protein, 3CLpro, and RdRp simultaneously so that basic proteins of SPs and NSPs could be inhibited and definitively the virus would be eradicated. It can be concluded that Linarin, Amentoflavone, (-)-Catechin Gallate, and Hypericin may have an effective role in SARS-CoV-2 infection. In addition to having a great affinity to attach to the viral proteins, these herbal compounds have antioxidant, vasoprotective, anticarcinogenic, and antiviral properties. Thus, they can be applied as extremely safe therapeutic natural compounds and clinical assessments might have notable outcomes for controlling COVID-19.
Acknowledgement :
The authors gratefully acknowledge AJMB Editorial office for English editing of this paper.
Conflict of Interest :
Authors declare no conflict of interest.
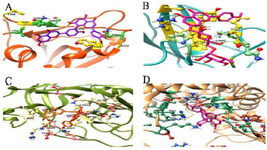
Figure 1. Affinity of top scoring medicinal herbal compounds to inhibit the S protein, 3CLpro, RdRp, and N protein. A) the interaction between Amentoflavone from Hypericum perforatum (Purple) and S protein (Orange red); B) the interaction between Linarin from Chrysanthemum morifolium (Deep pink) and N protein (Light sea green); C) the interaction between Amentoflavone from Hypericum perforatum (Orang) and 3CLpro (Olive drab); D) the interaction between Amentoflavone from Hypericum perforatum (Violet red) and RdRp (Sandy brown).
|

Table 1. Sixty-four traditional medicinal herbs used in the survey
|

Table 2. The binding affinity of top scoring chemical compounds for blocking each viral protein (N protein, 3CL, S protein, and RdRp) compared to available or proposed natural and antiviral drugs
|

Table 3. The binding affinity of the most common medicinal herbal compounds for each viral protein (N protein, 3CL, S protein, and RdRp)
|
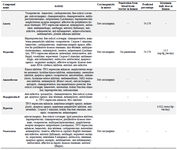
Table 4. Different characteristics of common medicinal herbal compounds having a high affinity to N protein, 3CL, S protein, and RdRp
|
|