Integrated Network and Gene Ontology Analysis Identifies Key Genes and Pathways for Coronary Artery Diseases
-
Prakash, Tejaswini
-
Genetics and Genomics Lab, Department of Studies in Genetics and Genomics, University of Mysore, Karnataka, India
-
 B Ramachandra, Nallur
Genetics and Genomics Lab, Department of Studies in Genetics and Genomics, University of Mysore, Karnataka, India, Tel: +91 9916344326, +91 9880033687; E-mail: prakash.tejaswini@gmail.com, nallurbr@gmail.com
B Ramachandra, Nallur
Genetics and Genomics Lab, Department of Studies in Genetics and Genomics, University of Mysore, Karnataka, India, Tel: +91 9916344326, +91 9880033687; E-mail: prakash.tejaswini@gmail.com, nallurbr@gmail.com
Abstract: Background: The prevalence of Coronary Artery Disease (CAD) in developing countries is on the rise, owing to rapidly changing lifestyle. Therefore, it is imperative that the underlying genetic and molecular mechanisms be understood to develop specific treatment strategies. Comprehensive disease network and Gene Ontology (GO) studies aid in prioritizing potential candidate genes for CAD and also give insights into gene function by establishing gene and disease pathway relationships.
Methods: In the present study, CAD-associated genes were collated from different data sources and protein-protein interaction network was constructed using STRING. Highly interconnected network clusters were inferred and GO analysis was performed.
Results: Interrelation between genes and pathways were analyzed on ClueGO and 38 candidates were identified from 1475 CAD-associated genes, which were significantly enriched in CAD-related pathways such as metabolism and regulation of lipid molecules, platelet activation, macrophage derived foam cell differentiation, and blood coagulation and fibrin clot formation.
Discussion: Integrated network and ontology analysis enables biomarker prioritization for common complex diseases such as CAD. Experimental validation and future studies on the prioritized genes may reveal valuable insights into CAD development mechanism and targeted treatment strategies.
Introduction :
Most common diseases that are manifested in human populations are complex in nature. Identifying the genes that cause these diseases has been challenging. However, the progress in recent years in high-throughput technology has led to the development of techniques that aid in gaining novel and important insights, resulting in a better understanding of the genetic architecture of complex diseases 1. Identification of genes and pathways involved in predisposing individuals to complex diseases is crucial to elucidate the pathophysiology of these diseases, which may eventually lead to the development of novel treatments 2. Coronary Artery Disease (CAD) is one such disease with complex interaction network and pathways which requires much attention for more specific and effective strategies of treatment.
CAD, a complex multifactorial disease, continues to be the leading cause of morbidity and mortality all over the world 3. India, in particular, has seen a steady raise in the number of CAD cases over the last 40 years 4. Several risk factors are known to contribute to CAD, such as, diabetes, hypertension, dyslipidemia, lifestyle and genetics 5. Over the years, multiple genetic studies have identified hundreds of causal and susceptibility loci associated with CAD that have improved our knowledge and understanding of its rudimentary causal factors 6-8. However, these studies have been successful in explaining only a small proportion of disease etiology 6, suggesting the involvement of many other genes in development of CAD that are yet to be defined. As the search for underlying genetic factors continues, biological validation of all the genes associated with CAD manifestation becomes difficult and impractical. Therefore, integration of high-throughput technology and bioinformatics has gained significant clinical relevance by improving the speed and efficiency of candidate gene discovery and prioritization 9. Leveraging these methods to understand the molecular mechanisms of CAD might improve our knowledge of biological relevance of CAD-associated genes, particularly in the context of complex networks 10.
In silico analyses using network and gene ontology-based approaches have been used widely to discover and prioritize potential candidate genes. The huge data generated from genome wide linkage and association or gene expression studies are extensively used to prioritize candidate genes and predict drug targets by means of network based approaches 11,12. The primary focus of network based approach is to discern the relationship between two components using Protein-Protein Interaction (PPI) patterns. A vast majority of research has gone into combining differential gene expression with network and pathway analysis to delineate the molecular mechanisms underlying CAD 10,13-17. Tan et al 10 used microarray data combined with network analysis to identify key genes and pathways in advanced coronary atherosclerosis. Similarly, Kashyap et al 18 established known candidates APOA1, CFTR, SRC, ICAM1, ESR1 and HNF1A as hub genes for CAD, Single Vessel Disease (SVD) and Triple Vessel Disease (TVD), respectively using PPI network and Gene Ontology (GO) analysis on differentially expressed genes in CAD subjects and normal controls. More recently, Miao et al integrated gene expression changes in CAD subjects with GO annotation and cluster analysis of PPI network, implicating IL1B, ICAM1, JUN and CCL2 in fluid shear stress, AGE-RAGE signaling pathway, Tumor Necrosis Factor (TNF) and cytokine-cytokine receptor interaction 19. There is also growing literature where studies have identified important CAD associated genes from existing literature and re-established their roles in CAD pathophysiology using PPI network-based approaches 4,13,17,20.
In view of these studies, an attempt was made to utilize phenotype association, PPI network, cluster analysis and GO annotation to identify highly interacting gene clusters and the interrelation between their biological functions to prioritize plausible candidates from CAD-associated genes.
Materials and Methods :
Selection of genes: Genes associated with CAD were retrieved from CardioGenBase (http://www.CardioGenBase.com/) 21. Collection of data from CardioGenBase was stopped on 26-03-2018. Additionally, Polysearch2 (http://pol-ysearch.ca/) 22 text mining tool was used to extract all associated genes/proteins given the disease term "coronary artery disease". The genes obtained from all these sources were combined and replicates were removed.
Disease association and phenotype enrichment analysis: Web-based Gene Analysis Toolkit (WebGestalt) 23 was used to perform disease association and phenotype enrichment analysis on the retrieved gene list with Homo sapiens as the organism of interest. Hypergeometric statistical test was used with Benjamini and Hochberg procedure to determine the false discovery rate. Significance level was set to "Top 10". Phenotype enrichment was established using Mammalian Phenotype Ontology and Human Phenotype Ontology. The genes enriched in phenotype analysis were further used for PPI analysis.
Protein-protein interaction network construction and module analysis: STRING v11.0 (Search Tool for the Retrieval of Interacting Genes/Proteins) (https://string-db.org/) 24 was used to construct PPI network using the gene set enriched in phenotype association analysis. Gene symbols were collated and duplicates were removed, retaining only non-redundant, unique genes. Homo sapiens was selected as the organism of interest. Experiments, databases and co-expression were used as active interaction sources, with a median confidence score of >0.4. Rest of the parameters were set to default. The resultant network was visualized in Cytoscape v3.8.0.0 25.
Module analysis was performed using Cytoscape's Molecular Complex Detection (MCODE) plug-in. MCODE identifies and clusters highly interconnected nodes in a network. PPI network was analyzed and clusters were inferred using the following parameters: node score cut-off - 0.2, node density cut-off - 0.1, K-core - 2, maximum depth - 100. Network scoring was done with a cut-off score of 2 and singly connected nodes were removed from clusters.
Gene ontology and pathway interrelation analysis of PPI network clusters: The genes identified from the top scoring clusters were subjected to function-based categorization to assess their functional association with biological processes, molecular function and cellular components with a significant p-value of <0.05. GO and pathway interrelation analysis of these genes was carried out on ClueGO, a Cytoscape plug-in, using annotations from KEGG. Right-sided hypergeometric test was employed to evaluate enrichment of genes in pathways with significant p-value (<0.05) corrected using Benjamini-Hochberg method.
Results :
Selection of genes: A total of 1441 genes were retrieved from CardioGenBase database. PolySearch text-mining source yielded 100 genes with Z-score>1. The two gene sets were combined to get a total of 1475 non-redundant, unique genes which were subjected to disease and phenotype association analysis.
Disease association and phenotype enrichment analysis: Disease association analysis performed on WebGestalt categorized the 1475 genes into 10 highly significant disease phenotypes -vascular diseases, cardiovascular diseases, myocardial ischemic disease, coronary disease, coronary artery disease, arterial occlusive diseases, arteriosclerosis, inflammation, heart diseases and genetic predisposition to diseases (Table 1).
Phenotype enrichment analysis showed the 1475 genes being enriched in three categories of phenotypic abnormalities; the interactions were represented as Directed Acyclic Graph (DAG)-abnormality of the integument, abnormality of blood and blood forming tissues (183 genes, adjP=9.83e-12), and abnormality of the cardiovascular system (Figure 1). The parent nodes (Shown in black) were further classified into nine significantly enriched phenotypic classes (Shown in red), with a collection of 253 genes.
Protein-protein interaction network construction and module analysis: PPI network was constructed on STRING database (Figure 2) using 253 seed genes collated from significantly enriched phenotypes. The resultant network had 158 nodes with 555 interactions. The network was analyzed for highly interconnected proteins using Cytoscape's MCODE plug-in and 13 clusters of connected nodes were obtained (Table 2), of which the top 3 high scoring clusters (Figures 3A and C) were selected for functional annotation. These clusters comprise 38 genes that can be potentially high-risk candidate genes for CAD.
Gene ontology and pathway interrelation analysis of PPI network clusters: GO analysis and pathway enrichment provided further insights into CAD related molecular mechanisms. The interrelation between pathways and 38 genes inferred from 3 high scoring network clusters was investigated using ClueGO (Figure 4). Significant enrichment was seen in GO terms such as regulation of plasma lipoprotein particle levels, regulation of lipid localization, regulation of lipid storage, plasma lipoprotein particle clearance, lipoprotein particle binding, plasma lipoprotein particle indicating that lipid metabolism and regulation plays a central role in CAD pathophysiology. Additionally, macrophage derived foam cell differentiation, platelet activation, complement and coagulation cascades, blood coagulation and fibrin clot formation, focal adhesion and AGE-RAGE signaling pathway in diabetes complications were also significantly enriched, giving insights into the pathophysiology of CAD. Detailed results of GO analysis are shown in table 3. The plausible biological function of each gene in relation to CAD is summarized in table 4.
Discussion :
The major underlying cause for CAD is Atherosclerosis (AS), which is developed from a complicated etiology that involves complex interactions between several genes and proteins 26. The treatment strategies for AS-related diseases are currently limited to the use of anticoagulant and lipid-lowering drugs. Therefore, furthering our knowledge of AS mechanisms and identifying novel targets for therapeutics holds significant clinical value. Although Genome-Wide Association Studies (GWAS) and the more recent high throughput technologies such as, Next Generation Sequencing (NGS) and genome expression profiling have established association of hundreds of probable candidate genes to CAD, identifying specific disease-causing genes and the mechanism through which they contribute to disease progression has been challenging. Also, experimental validation of vast number of genes is time consuming and laborious 4. Systems biology offers a feasible approach for prioritization of genes and elucidation of their roles in manifestation of disease. Genes and proteins that are most relevant to disease can be mined and analyzed from large public databases using in silico analyses. Thus, it is deemed to be a promising technology to discover novel targets for treatment.
Recent years have seen remarkable progress in biomarker identification for CAD using network-based approaches. Genes associated with CAD and related phenotypes have been used to construct PPI networks and identify selected hub genes that are possibly most relevant to disease manifestation and progression. In the present study, an integrated approach of phenotype association, PPI network and gene ontology analysis was employed to identify 38 plausible high-risk candidate genes from 1475 CAD-associated genes. GO analysis and pathway enrichment analysis of 38 candidate genes demonstrated several of them are established CAD driver genes (AGXT, APOA1, APOB, APOE, LCAT, LDLR, LIPC, LPA and LPL) and participate in the lipid metabolic processes. Significant enrichment was observed in macrophage derived foam cell differentiation (AGXT, APOB, ITGB3 and LPL). Macrophages are critical players in the development of atherosclerosis as they contribute to plaque development, regulate and maintain vascular inflammatory response, and stimulate thrombus formation, making them desirable therapeutic targets 27. Blood coagulation and fibrin clot formation (F10, F11, F12, F2, F9, FGA, GP1BA, GP9) and regulation of wound healing process (APOE, F11, F12, F2, FGA, GP1BA, HRAS) were also significantly enriched. GO annotation also showed enrich-ment of genes in Extracellular Matrix (ECM) structural constituents which play crucial roles in cell integrity, adhesion and communication. ECM governs the proper functioning of different organs and changes in its components have been implicated in CAD mechanisms such as atherothrombosis, plaque rupture and calcification. Vascular smooth muscle cells and macrophages functioning are also shown to be influenced by matrix constituents such as collagens 28. Previous network analysis by Zhao et al 16 has also established the importance of ECM proteins in CAD development and its link with complement and coagulation cascades, and inflammation.
The study also identified less well-studied possible candidates for CAD in addition to established CAD driver genes such as, APP and FBN1. Amyloid Precursor Protein (APP) encodes amyloid-β (Aβ) peptides which are main drivers of Alzheimer's Disease (AD), a progressive neurodegenerative disorder. The biological function of APP with respect to Alzheimer's disease has been scrutinized extensively over the last 30 years 29. Interestingly, in addition to being expressed in human brain, kidney, and platelets, APP is also expressed in vascular endothelium of coronary, cerebral and peripheral blood vessels. Additionally, the endothelial cells of large conduit arteries, resistance arteries, and microvessels also show the expression and activity of enzymes responsible for proteolytic cleavage of APP 29. In a study of apolipoprotein E knockout (ApoE-/-) mice, Tibolla et al 30 and, Austin and Combs 31 demonstrated accelerated aortic atherosclerotic development and endothelial dysfunction induced by APP overexpression. Austin and Combs also went on to show that in ApoE-/- mice and AD patients, there was overexpression of APP mediated monocyte adhesion to the endothelium, implying that vascular dysfunction associated with AD and atherosclerosis involves endothelial APP function. However, the molecular mechanisms and physiological outcomes underlying APP action are incompletely understood, thereby making it an interesting candidate for future study. Similarly, the effect of impaired FBN1 action was studied using mouse models by Van der Donckt et al 32. FBN1 mutations have been established to cause Marfan syndrome, affecting the connective tissues that support the body’s joints and organs. Van der Donckt et al 32 showed that C1039G+/− mutation in mice resulted in increased arterial stiffness owing to fragmentation of the elastic fibers of the vessel wall. It was also seen that extremely unstable plaques were developed due to the mutation leading to sporadic ruptures. Although several single nucleotide polymorphisms and mutations have been mapped to FBN1, the precise mechanistic action through which it exerts arterial stiffness and aids atherosclerotic progression has not been established.
Pathway enrichment analysis performed on the prioritized genes asserted some results of previous studies. Over-representation of genes was seen in AGE-RAGE signaling pathway in diabetes complications. Diabetes Mellitus (DM) is a well-established risk factor for CAD and is known to promote vascular calcification through mechanisms of hyperglycemia, hypercalcemia and oxidative stress. Several studies have implicated the AGE/RAGE signaling pathway in accelerating vascular calcification in DM patients which increases the risk of atherosclerotic plaque rupture, resulting in the possibility of heart attack or stroke 33-35. AGE-RAGE signaling pathway is also a key component of inflammation and immune response. Miao et al 19 identified 413 differentially expressed genes using two datasets of gene expression studies in CAD subjects and controls and emphasized the contribution of AGE-RAGE signaling pathways in the pathogenesis of CAD. Significant enrichment was also observed in focal adhesion, platelet activation pathways and complement-coagulation cascades. Focal adhesion molecules are important mediators of interaction between blood vessels and circulating leukocytes 36. They control adhesion dynamics and influence the interaction between platelets and endothelium. They also play a key role in vascular dysfunction and tissue injury in atherosclerosis 37. An integrative network and pathway analysis by Chan et al 38 showed that the CAD and diabetes molecular pathways share focal adhesion as one of the key pathways in different ethnicities. Several key processes involved in the development and progression of atherosclerosis are shown to be influenced by the complement system through in vitro studies. It is also said to influence the extent of thrombus formation when the atherosclerotic plaque ruptures 39. As demonstrated by the pathway interrelation analysis, these pathways function with a complex network of interconnected genes. Therefore, dysfunction or disruption of these important genes may result in a chain of functional pathological outcomes.
Conclusion :
The current study focused on prioritizing CAD-associated genes and identified 38 potential high-risk CAD candidates using a combination of network and pathway analysis in a stepwise manner. These genes were found to be involved in key biological processes and pathways that contribute to the pathogenesis of CAD. Therefore, it can be concluded that if any of these genes in the network fails to function normally owing to variations and/or mutations, the downstream processes can be disrupted resulting in development of CAD. The strength of the study lies in ontology-based gene filtration. However, there are certain limitations to this study. Since the knowledge of CAD genetics and mechanisms is still an ongoing process, network construction could be incomplete or certain interactions could be missing as it is derived from published reports and these results need to be further validated with appropriate wet-lab experiments to delineate the biological mechanisms.
Acknowledgement :
We are grateful to Department of Studies in Genetics and Genomics, University of Mysore for providing facility to conduct this work. We also thank members of the Genetics and Genomics lab, Department of Studies in Genetics and Genomics for their support and encouragement. We thank Institution of Excellence for providing fellowship to Ms. Tejaswini Prakash.
Conflict of Interest :
The authors declare no competing financial interests.
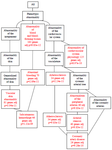
Figure 1. Directed acyclic graph showing phenotype enrichment of 1475 CAD-associated genes. Nodes in black represent non-enriched parent nodes. Nodes in red represent significantly enriched phenotypes.
|
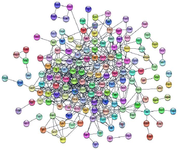
Figure 2. Protein-protein interaction network constructed from genes derived from phenotype enrichment analysis.
|
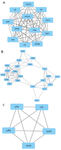
Figure 3. A-C) Top scoring network clusters from module analysis using Cytoscape's MCODE plug-in.
|
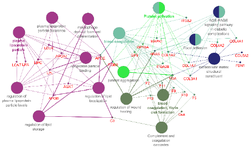
Figure 4. Pathway interrelation analysis of genes derived from top scoring network clusters.
|
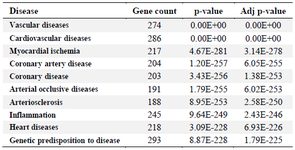
Table 1. Disease association analysis of CAD-associated genes showing top 10 significantly enriched disease categories
|
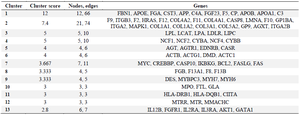
Table 2. Highly interconnected genes in network clusters derived from module analysis using Cytoscape's MCODE plug-in
|
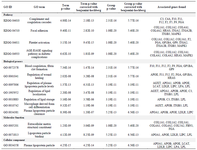
Table 3. Gene ontology and pathway enrichment analysis of 38 identified candidate genes. Enrichment is classified into pathways, biological processes, molecular functions and cellular components
|

Table 4. Plausible function of 38 prioritized candidate genes for CAD
|
|