Construction and Production of Foxp3- Fc (IgG) DNA Vaccine/Fusion Protein
-
Mousavi Niri, Neda
-
Department of Medical Biotechnology, Faculty of Advanced Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran
-
 Hadjati, Jamshid
Department of Immunology, Faculty of Medicine, Tehran University of Medical Sciences, Tehran, Iran, Tel: +98 41 33355788, E-mail: zarghami@tbzmed.ac.ir
Hadjati, Jamshid
Department of Immunology, Faculty of Medicine, Tehran University of Medical Sciences, Tehran, Iran, Tel: +98 41 33355788, E-mail: zarghami@tbzmed.ac.ir
-
Department of Immunology, Faculty of Medicine, Tehran University of Medical Sciences, Tehran, Iran
-
Shokri, Mehdi
-
Department of Hepatitis and AIDS, Pasteur Institute of Iran, Tehran, Iran
-
Pilehvar-soltanahmadi, Yones
-
Department of Medical Biotechnology, Faculty of Advanced Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran
-
Akbarzadeh, Abolfazl
-
Department of Medical Nanotechnology, Faculty of Advanced Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran
-
 Zarghami, Nosratollah
Department of Medical Biotechnology, Faculty of Advanced Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran. Department of Clinical Biochemistry, Radiopharmacy Lab, Drug Applied Research Center, Tabriz University of Medical Sciences, Tel: +98 41 33355788, E-mail: zarghami@tbzmed.ac.ir
Zarghami, Nosratollah
Department of Medical Biotechnology, Faculty of Advanced Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran. Department of Clinical Biochemistry, Radiopharmacy Lab, Drug Applied Research Center, Tabriz University of Medical Sciences, Tel: +98 41 33355788, E-mail: zarghami@tbzmed.ac.ir
-
Department of Medical Biotechnology, Faculty of Advanced Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran
-
Department of Clinical Biochemistry, Radiopharmacy Lab, Drug Applied Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
Abstract: Background: It seems that the success of vaccination for cancer immunotherapy such as Dendritic Cell (DC) based cancer vaccine is hindered through a powerful network of immune system suppressive elements in which regulatory T cell is the common factor. Foxp3 transcription factor is the most specific marker of regulatory T cells. In different studies, targeting an immune response against regulatory cells expressing Foxp3 and their removal have been assessed. As these previous studies could not efficiently conquer the suppressive effect of regulatory cells by their partial elimination, an attempt was made to search for constructing more effective vaccines against regulatory T cells by which to improve the effect of combined means of immunotherapy in cancer. In this study, a DNA vaccine and its respective protein were constructed in which Foxp3 fused to Fc(IgG) can be efficiently captured and processed by DC via receptor mediated endocytosis and presented to MHCII and I (cross priming).
Methods: DNA construct containing fragment C (Fc) portion of IgG fused to Foxp3 was designed. DNA construct was transfected into HEK cells to investigate its expression through fluorescent microscopy and flow cytometry. Its specific expression was also assessed by western blot. For producing recombinant protein, FOXP3-Fc fusion construct was inserted into pET21a vector and consequently, Escherichia coli (E. coli) strain BL21 was selected as host cells. The expression of recombinant fusion protein was assayed by western blot analysis. Afterward, fusion protein was purified by SDS PAGE reverse staining.
Results: The expression analysis of DNA construct by flow cytometry and fluorescent microscopy showed that this construct was successfully expressed in eukaryotic cells. Moreover, the Foxp3-Fc expression was confirmed by SDS-PAGE followed by western blot analysis. Additionally, the presence of fusion protein was shown by specific antibody after purification.
Conclusion: Due to successful expression of Foxp3-Fc (IgG), it would be expected to develop vaccines in tumor therapies for removal of regulatory cells as a strategy for increasing the efficiency of other immunotherapy means.
Introduction :
The immune system exploits a network of central and peripheral tolerance mechanisms to discriminate between self and non-self. One of the main components of this network is CD4+CD25+regulatory T cells (T reg) whose function is to suppress immune responses 1. As well as CD25, other markers such as GITR, CTLA-4, CD103 and OX-40 are overexpressed on T reg cells but their expression is not as specific as CD25 on regulatory T cells 2. A transcription factor called FoxP3, a member of the fork head family of transcription factors, is specifically expressed by T regs. Regulatory T cells control the immune response either by their direct contact with the immune cells or with the secretion of soluble factors. The importance of regulatory cells in maintaining self-tolerance is illustrated by deficiency in their content and function in many autoimmune diseases 3. Evidences from mouse model systems and cancer patients indicate that regulatory T cells affect anti-tumor responses in tumor-bearing individuals 2-7. Two sets of observations also implicate T reg in suppression of tumor immunity. First, the numbers of T reg increased at the tumor sites of cancer patients correlated with disease progression. Second, depletion of regulatory T cells in mice enhances antitumor immunity and reduces tumor growth 3. In spite of that, in recent clinical trial, elimination of T reg in renal cancer patients using an interleukin 2(IL2)/diphtheria toxin fusion product (ONTAK) led to enhanced vaccine-induced anti-tumor immune responses. Therefore, elimination of T reg could represent an important adjunct to cancer immunotherapy 8.
The only gene product known to be exclusively expressed in T reg of mice is FoxP3. FoxP3 is an intracellular factor which its expression is not only in CD4+CD25+T cells but also in CD4+CD25low/- T reg, as well as, subsets of CD8+T cells that exhibit immune-suppressive properties. Thus, targeting FoxP3 offers distinct advantages over targeting CD25 to eliminate immune suppressive cells in vivo 3. Since FoxP3 is an intracellular product, FoxP3 expressing T regs cannot be destroyed using monoclonal antibodies 9. CD8+Cytotoxic T cells (CTL) could recognize cellular products combined with MHCI on cell surface. FoxP3 laboring cells could be targeted by CTL in the same way 10.
Generally, different means have been exploited in T reg suppression by researchers such as chemical drugs 9,11,12, anti CD25 monoclonal anti body 13,14, immunotoxins [Denileukin diftitox (ONTAK) and LMB-2 (single chain fragment variable anti-tacfused with bacterial Pseudomonas exotoxin A)] 1,8,15 and anti T reg vaccination targeting FoxP3 3. To date, there have been several strategies in targeting FoxP3 for T reg suppression 3,9,16. In a project by Generali et al, T regs were modulated by letrozole which is one of Aromatase inhibitors and impairs FoxP3 signaling 9. In 2007, Nair et al showed that depletion of regulatory T cells using dendritic cells pulsed with mRNA of FoxP3 could enhance effect of therapeutic anticancer vaccination 3. Overall, depletion of T regs in transgenic manner also improves therapeutic anticancer immune properties of effector cells 17.
Antigen immunogenicity can be augmented in their fusion with fragment C (Fc) of immunoglobulin heavy chain leading to antigen-Fc fusion protein. The antigen-Fc fusion protein attaches to Fc receptors on the surface of antigen expressing cells (APCs) and antigen can be targeted by these cells in mammalian cells 18. In some researches, fusion of fragment C of immunoglobulin G (IgG) to different antigens such as tumor antigens could stimulate higher immune responses compared to antigens alone 19. You et al showed that fusion of hepatitis B antigen to Fc (IgG) in a DNA vaccine format led to enhanced capture and presentation of antigen by dendritic cell. The respective fusion protein produced by this DNA vaccine could induce B cell response more effectively. As well as its efficient receptor-mediated endocytosis by dendritic cell, it could also be better presented on MHCI and MHCII. Totally, the antigen-Fc fusion caused considerable increase in antigen specific responses of CD4+T cell, CD8+CTL and B cell 20. Apart from enhancing the antigenic stimulation, Ig(Fc) fusion has been shown to possess other advantages, too. Chemokine/cytokine-Ig fusion presents the advantages of divalent affinity, non-cytolytic effect and long in vivo half-life with conserved activity of both proteins 21,22. The main objective of this study was cloning and expression of recombinant vectors containing FoxP3-IgG2Fc with the purpose of DNA vaccine and recombinant protein production (As prime/boost vaccination regimen in future studies) by a simple one step procedure and evaluation of their proper work ex vivo and in vitro, respectively.
Materials and Methods :
Plasmids and bacterial strains: pEGFPN1-FoxP3 and pET24a-FoxP3 plasmids which were previously constructed by our research group were truncated FoxP3 genes cloned in pEGFPN1 and pET24a vectors, respectively. Truncated FoxP3 lacks a polypeptide segment called nuclear localization signal and its shortage leads to impaired functional properties of FoxP3. pIRES2-EGFP-IL18-Fc(IgG) was a gift from another research group 22. Escherichia coli (E. coli) strains, DH5α and BL21 (DE3), and plasmids, pIRES2-EGFP and pET21a, were obtained from National Recombinant Gene Bank (NRGB), Pasteur Institute of Iran.
Bacterial cultures: The E. coli strains were grown in LB broth (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl, pH=7.0) and on LB agar with Kanamycin and Ampicilin (Sigma).
Chemicals and enzymes: IPTG, T4 DNA ligase and Pfu DNA polymerase were purchased from Fermentase (Lithuania). Chemicals were obtained from Merck (Germany). Restriction endonucleases were purchased from Enzynomics (Korea). PolyFect transfection kit was obtained from Qiagen (Germany).
Gene amplification and cloning procedures: Truncated (1114 bp) FoxP3 fragment (correspond-ing to amino acids 1-363) was created by PCR amplification on previously constructed pEGFP-N1-FoxP3 plasmid, using NheI-tailed forward (5ˊATAT GCTAG CGCCACCATGGCTC3ˊ), BglII-tailed reverse (5ˊTA CAGATCTGGCGAACATGCGAG3ˊ) primer pairs and proofreader Pfu DNA polymerase (Thermo, USA) in a thermal program of 94°C (4 min) and 30 cycles of 94ºC (40 s), 55ºC (40 s) and 72ºC (68 s). The kozak Seq (ACCATGG) was already included in the upstream of truncated FoxP3 gene in pEGFP-N1-FoxP3 plasmid that was used as a template for PCR cloning of FoxP3 into pIRES plasmid. Also, kozak sequence was in our designed NheI-tailed forward primer. As it is shown in figure 1A, to construct pIRES2-EGFP-FoxP3-IgG2Fc plasmid that encoded the fusion of truncated FoxP3-IgG2Fc, amplified FoxP3 truncated gene was double digested with NheI/BglII enzymes and further replaced with Igk-IL18 segment in previously constructed pIRES2-EGFP-Igk-IL18-IgG2Fc plasmid. This plasmid expresses FoxP3-IgGFc fusion protein and additionally encodes for EGFP as a separate protein.
Additionally, pET21-FoxP3-IgG2Fc plasmid was constructed by PCR amplification on previously constructed pIRES2-EGFP-FoxP3-IgG2Fc plasmid, using EcoRI-tailed forward (5ˊTGGAATTCGCTCCTTCCT TGG3ˊ), HindIII-tailed reverse (TATAAGCTTTAG CCCCGGAGTCC) primer pairs and proofreader Pfu DNA polymerase (Thermo, USA) in a thermal program of 94°C (4 min) and 30 cycles of 94ºC (40 s), 56C (40 s) and 72ºC (240 s). EcoRI-FoxP3-IgG2Fc-HindIII fragment was cloned into pET21a to construct pET21-FoxP3-IgG2Fc plasmid. This plasmid encodes for T7tag-FoxP3-IgG2Fc fusion protein (Figure 1B).
Finally, the precision of cloned genes in all recombinant plasmids of pIRES2-EGFP-FoxP3-IgG2Fc and pET21-FoxP3-IgG2Fc was checked and confirmed by restriction endonuclease double digestion and commercially available sequencing services (Sequencing Lab., Pasteur Institute of Iran).
Cell line, transfection, and preparation of cell lysate: HEK-293 cell line was maintained in Dulbecco’s Modified Eagle Medium (DMEM, 1x) containing 2.0 mM L-gluthamin, 100 U/ml penicillin,100 mg/ml streptomycin and 10% Fetal Bovine Serum (FBS) at 37oC, 5% CO2. Cells were transiently transfected with the DNA construct PIRES-GFP-Foxp3-IgG2Fc by polyfect transfection reagent (Qiagen, Germany). In brief, the transfection complex was prepared according to the optimized amount of plasmid and polyfect reagent mentioned in manufacturer instruction and transferred to 80% confluent HEK-293 cells.
At 72 hr post-transfection, transfected cells were either assessed for fluorescence microscopy analysis and flowcytometry or subjected to lysis with the mixture of 0.1 M Tris-Cl (pH=7.8) and 0.5% (V/V) Triton X-100.
Gene expression assays
Fluorescent microscopy and flowcytometry analysis: At 72 hr post-transfection, the flourescence of transfected cells was analyzed with a Zeiss Axioskop fluorescence microscope and non-transfected cells were used as the negative control.
At the same time, trypsinized cells were analyzed for GFP emission after gating on live population by means of Partec (PAS) cytometer instrument and FlowMax software (Partec, Germany).
Western-blotting: Cell lysates were separated in 12% SDS-PAGE under reducing condition, transferred to 0.45 µm pore size polyvinylidene difluoride (PVDF) membrane (Hi-bond Amersham Biosciences, USA) by using a semi-dry blotter unit (Biorad, USA) and blocked by 5% Bovine Serum Albumin (BSA). Subsequently, membrane was incubated with goat anti-mouse immunoglobulin G (heavy and light chain) Horseradish Peroxidase (HRP) conjugate antibody (Sigma, USA) for one hr at room temperature. The antibody was diluted 1:5000 in BSA. Detection of the protein was achieved with 3,3-diaminobenzidine tetrahydrochloride (DAB) reagent (Sigma, Saint Louis, MO, USA) and placement in darkness. Lysate of non-transfected cells served as the negative control.
The similar steps were done to confirm the expression of recombinant FoxP3-IgG2Fc fusion protein in bacterial host as well.
Expression and purification of Foxp3-IgG2Fc recombinant protein: To express the recombinant Foxp3-Fc(IgG) fusion protein, pET21-FoxP3-IgG2Fc plasmid was transformed into competent E. coli BL21(DE3) and transformed cells were grown at 37ºC in LB medium supplemented with 50 µg/ml ampicillin (MERCK) up to exponential phase (OD600 nm=0.5), followed by induction of 1 mM IPTG. Samples were collected 3 hr, 5 hr and overnight post induction and the cell pellets were analyzed by SDS-PAGE to follow the best time point of protein expression. Electrophoresis of protein samples was followed by floating the gel in sodium carbonate solution (0.08 M). Then the gel was pretreated in imidazole-SDS solution [200 mM imidazole, 0.1% (m/V) SDS followed by developing in 200 mM zinc sulfate] until the gel background turned intensely white with transparent protein bands. As development of background continued for a few seconds after the developer was discarded, the reaction was best stopped just as the bands of interest became visible. Then the band of interest was cut and homogenized in protein extraction buffer (ammonium carbonate and SDS) and was agitated overnight on the rocker. The suspension was centrifuged (20 min, 4ºC, 3800 rpm) and the supernatant was collected to be concentrated by Viva spin concentrator tubes (Sartorius, Germany) with 3 kd cut off. Protein concentration was determined by Nano drop analyzer (Thermo scientific, nanodrop1000 spectrophotometer, USA) and the purity was determined by SDS-PAGE and Coomassie Brilliant Blue (R-250) staining.
Results :
Construction of DNA plasmids that encode truncated FoxP3-IgG2Fc fusion: The truncated Foxp3 fragment was amplified by PCR on pEGFP-N1-FoxP3 using specific primers. A 1% agarose gel was run to confirm the existence of proper size of PCR product and only a single band was appeared with an estimated size of ~1114 base pairs (Figure 2). The double digested PCR product NheI-FoxP3-BglII was cloned into pIRES2-EGFP-Igk-IL18-IgG2Fc plasmid to replace IL18 with FoxP3 and made pIRES2-EGFP-FoxP3-IgG2Fc plasmid with total size of 7058 bps (Figure 1A). Then the construct was subsequently transformed into competent E. coli DH5α cells. The resultant colonies were evaluated for the true insert size by colony PCR method, two different enzymatic digestions (Figure 3) and PCR on colony extracted plasmids.
PCR reaction on pIRES2-EGFP-FoxP3-IgG2Fc plasmid using F2/R2 primer pair created the FoxP3-IgG2Fc fragment with size of 1830 bps (Figure 4A). Then the double digested PCR products and Pet21a vector were ligated together (Figure 1B) at a molar ratio of 6:1 and subsequently transformed into competent E. coli DH5α cells. The resultant colonies were evaluated with the same methods as previous ones. Figure 4B indicates enzymatic digestion of pET21a-Foxp3-IgG2Fc construct with two restriction enzymes EcoRI and HindIII. In the next step, recombinant vector, pet21a-Foxp3-IgG2Fc (Figure 4C), was applied for the transformation of E. coli BL21 (DE3) cells. The accuracy of FoxP3-Fc fusion gene in pIRES2-EGFP-FoxP3-IgG2Fc and pET21a-Foxp3-IgG2Fc plasmids was confirmed by sequencing reactions (Data not shown).
Eukaryotic expression of recombinant FoxP3-IgG2Fc: The pIRES2-EGFP-FoxP3-IgG2Fc plasmid expressed FoxP3-IgGFc fusion protein and additionally was encoded for GFP as a separate protein. Fluorescence microscopy of transfected HEK 293 cells versus non-transfected ones roughly showed the expression of GFP protein (Figures 5A and 5B). Furthermore, flow cytometric analysis quantified GFP-emitting cells as 40% of the live population (Figures 5C and 5D) with mean fluorescence intensity (MFI) of 29 (for GFP positive population) versus 0.6 (for GFP negative population). This EGFP emission at least confirmed the successful transcription of GFP-FoxP3-IgG2Fc mRNA, which is an indirect indicator for FoxP3-IgG2Fc expression. To have a direct assessment of protein expression, lysates of HEK 293 cells transfected with pIRES2-EGFP-FoxP3-IgG2Fc plasmid were analyzed by western blotting. Results indicated the ex vivo expression of the FoxP3-IgG2Fc fusion protein with the expected size of around 69 kDa in HEK 293 cells (Figure 5D). These data confirm that pIRES-EGFP-FoxP3-IgG2Fc plasmid properly expresses the FoxP3-IgG2Fc protein ex vivo, and hence, is qualified to be tested as a DNA vaccine plasmid for in vivo expression of this fusion protein.
Prokaryotic expression and purification of recombinant Foxp3-IgG2Fc fusion protein: E. coli BL21 (DE3) cells that were transformed with pET21a-FoxP3-IgG2Fc construct were induced for protein expression by adding IPTG. Lysates from induced and non-induced cells were then separated using SDS-PAGE. After staining of proteins, a band with an approximate size of ~69 kDa on lane of the induced samples was expected to be the protein of interest (Figure 6A).
In order to determine which time course of induction resulted in a higher protein production level, the same amount of IPTG, 1 mM, was added to cultures (OD 600 0.4-0.6) and at different time points of 3 hr, 5 hr and overnight post-induction, production levels of target protein were quantified. The optimal amount of rFoxp3-IgG2Fc was attained over the period of 3hrs after induction with IPTG (Figure 6B).
Purification and confirmation of recombinant Foxp3- IgG2Fc: Therefore, the high expression level of rFoxp3-IgG2Fc was obtained when rFoxp3- IgG2Fc expressing E. coli BL21(DE3) cells were grown at 37oC in LB broth supplemented with 50 μg/ml ampicillin and induction of cells were conducted with 1 mM IPTG after the OD 600 nm reached 0.6. The optimal expression level was found to be achieved 3 hr after the addition of IPTG to the medium. Recombinant protein was purified by Imidazole-SDS-Zinc reverse staining. Theoretical molecular weight of target protein was measured ~69 kDa. The 69 kDa protein band was observed in SDS-PAGE and it was confirmed as rFoxp3- IgG2Fc protein by western blot analysis using goat anti-mouse antibody (Figures 7A and 7B).
Overall, these data confirm that pET21-FoxP3-IgG2Fc plasmid can be efficiently used in an optimized system for preparative expression and purification of truncated FoxP3-IgG2Fc protein for further aims of animal immunization and vaccine studies.
Sequence analysis of recombinant Foxp3- IgG2Fc: The final nucleotide sequence for constructed FoxP3-IgG2Fc was 98% matched with the expected nucleotide sequence. However, the amino acid translation of this sequence was completely (100%) matched with the expected amino acid sequence for truncated FoxP3-IgG2Fc protein (Data not shown).
Discussion :
The application of DNA vaccine and its respective recombinant protein production in biotechnology have been increased in experimental and clinical designs. However, high efficiency of DNA vaccines and recombinant proteins in experimental systems is an issue. Therefore, different methods including fusion with different partners have been developed to improve their effectiveness.
Due to their rapid and widespread development, DNA vaccines have entered into a variety of human clinical trials for vaccination against various diseases including cancer. The results of previous studies in clinical trial have shown that such DNA vaccines require much improvement in antigen expression and delivery methods to make them sufficiently effective in the clinic. Similarly, additional strategies should be employed to activate effective immunity against poorly immunogenic tumor antigens. Engineering vaccine design for manipulating antigen presentation and processing pathways is one of the most important aspects that can be easily handled in DNA vaccine technology 23. Several approaches have been investigated including DNA vaccine engineering, co-delivery of immuno-modulatory molecules, safe routes of administration, prime-boost regimen and strategies to break the immunosuppressive network mechanisms adopted by malignant cells to prevent immune cell function 24. Combined or single strategies to enhance the efficacy and immunogenicity of DNA vaccines are applied in completed and ongoing clinical trials, where the safety and tolerability of the DNA platform are substantiated 23-26.
APCs are critical for initiating and modulating B- and T-cell responses elicited by DNA vaccination. However, only a very limited fraction of injected DNA molecules are taken up by APCs in draining lymph nodes 27. Even when DCs are transfected, the intracellular antigens expressed by DCs are difficult to be processed and presented to MHC class II 10. Even secretory antigens cannot be efficiently presented to MHC class I and II because of the inefficiency of internalization of soluble antigens through fluid phase pinocytosis 20. In this study, antigen-Fc fusion protein was constructed which aids receptor-mediated internalization process to enhance APC antigen presentation and increase immune responses, especially CD4+T cells. Although there have been attempts to target an antigen to APCs to enhance the potency of DNA vaccines, such as using Cytotoxic T-Lymphocyte-Associated protein 4 (CTLA4) molecule 28,29, the vaccine design described has some unique features. Fc portion enables receptor-mediated endocytosis pathway, permits fusion antigens to be efficiently captured, processed in endosomes, and presented to MHC by APC to induce T cell responses 20.
As described in this project, DNA construct containing Fc portion of IgG fused to Foxp3 was designed in pIRES-EGFP plasmid. The expression of DNA construct was investigated by transfection of HEK cells with this DNA construct.
It has been reported that combination of tumor antigen expressing vector and plasmids containing other proteins such as adjuvants (to improve immune responses) could ameliorate antitumor reactions in vivo 24. Moreover, some reports have shown that presence of bicistronic vectors is also helpful for efficiency of gene adjuvants 30. One of the characteristics of these vectors is production of single long mRNA from two different expressing regions under the control of a single promoter. In this mRNA, the first protein is translated under CAP-dependent mechanism and the second is produced using IRES 30. In some researches on positive role of adjuvants in improved induction of immune response, it has been shown that expression of both tumor antigen and adjuvant by single plasmid could cause more powerful immune responses 31. In other words, a good stimulation of lymphocytes is following co-presentation of antigen and adjuvant by antigen presenting cells 32. Using adjuvants with tumor antigens in combination with distinguished immunotherapy means is considered as a combinatorial therapy 33.
FoxP3-IgG2Fc fusion construct and protein produced in this study targets regulatory T cells which are an obstacle against efficient anti-tumor responses. Therefore, combination of the present FoxP3-IgG2Fc fusion system and other conventional immunotherapies seems encouraging to attain effective, reliable and consistent clinical efficacy. In the present study, truncated FoxP3 gene fused to IgG2Fc was amplified and cloned into pIRES2-EGFP and pET21a vectors successfully. Subsequently, ex vivo performance of both vectors was checked in prokaryotic and eukaryotic expression systems, respectively. Thus, it is required to go further to test our constructs in vivo and see whether they can induce a functional immune response against T regs.
Conclusion :
This work was supported by Department of Biotechnology, Faculty of Advanced Medical Sciences, Tabriz University of Medical Science, Department of Hepatitis and AIDS, Pasteur Institute of Iran, Department of Immunology, Faculty of Medicine, Tehran University of Medical Science and Iran National Science Foundation (INSF).
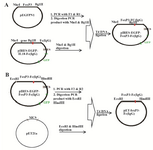
Figure 1. Cloning strategies for constructing the A) pIRES2-EGFP-FoxP3-IgG2Fc and B) pET21a-FoxP3-IgG2Fc vectors.
|
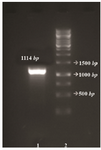
Figure 2. Agarose gel electrophoresis of the PCR amplified fragment. From left to right, Lane 1: negative control, Lane 2: PCR product, Lane3: DNA marker 1 Kb (Fermentase).
|
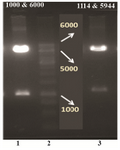
Figure 3. Restriction analysis of the pIRES2-EGFP-FoxP3-IgG2Fc construct with two different enzymatic cocktails. Lane 2: DNA marker 1 Kb (Fermentase), Lane 1 and 3: digested forms of the recombinant plasmid by HindIII/BglII and NheI/BglII enzymatic cocktail, respectively.
|
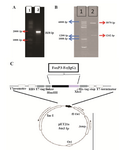
Figure 4. A) Agarose gel electrophoresis of the PCR amplified frag-ment. From left to right, Lane1: DNA marker 1 Kb (Fermentase), Lane 2: PCR product; B) restriction analysis of the pET21a-Foxp3-IgG2 (Fc) construct. From left to right, Lane 1: DNA marker 100 bp (Fermentase), Lane 2: digested form of the recombinant plasmid by BglII enzyme; C) schematic representation of the expression elements in the pET21a-Foxp3-IgG2(Fc) plasmid. TheFoxp3-IgG2(Fc) nucleotide sequence was ligated into the BglII/HindIII sites of the pET21a plasmid. This cloning strategy permitted to fuse highly ef-ficient Ribosome Binding Site from the phage T7major capsid protein and T7 tag to the N-terminal and a6xHis tag to the C-terminal of the Foxp3-IgG2 (Fc) fragment.
|
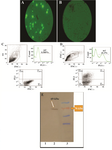
Figure 5. A) Evaluation of transient transfection of pIRES2-EGFP-FoxP3-IgG2Fc in HEK 293T cells compared to B) non-transfected (negative control) using fluorescent microscopy (100X) and C, D) dot-plot and histogram views of flow cytometric analysis for GFP expression in non-transfected (control) compared to transfected cells, respectively; E) assessment of the expression for FoxP3-IgG2 (Fc) fusion protein in cell lysates of pIRES2-EGFP-FoxP3-IgG2Fc transfected HEK 293T cells was carried out by using anti-Fc(IgG) polyclonal antibody. Lane 1: negative control, Lane 2: the band for fusion protein of interest with the size of 69 kDa (indicated arrow), Lane 3: protein MW marker.
|
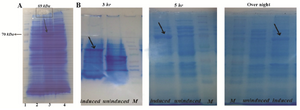
Figure 6. A) SDS-PAGE analysis of recombinant Foxp3-IgG2 (Fc) with Coomassie-stained. Expression of rFoxp3-IgG2 (Fc) in E coli BL21 induced with 1.0 mmol IPTG. From left to right: Lane 1: Protein marker (Fermentase), Lane 2: Non-induced with IPTG, Lane 3: Induced with IPTG (~69 kDa), Lane 4: Bacterial lysate (negative control); B) the effect of the period of induction on high production level of rFoxp3- IgG2(Fc). Triplicate lanes induced with 1mM IPTG (showed by arrows), uninduced and protein marker at time course of induction 3 hr, 5 hr and overnight, respectively.
|
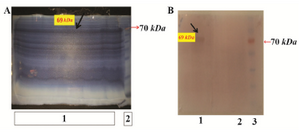
Figure 7. A) Purification of rFoxp3- IgG2 (Fc) by Imidazole-SDS-Zinc reverse staining. From left to right: Lane 1: transformed bacterial lysate induced by IPTG which was run in a wide lane and shown by arrow, Lane 2: protein marker; B) western blot analysis. From left to right, Lane 1: induced E. coli BL21containing Pet21a-FoxP3-IgG2(Fc), Lane 2: negative control, Lane 3: protein marker.
|
|