Inhibition of Coenzyme Qs Accumulation in Engineered Escherichia coli by High Concentration of Farnesyl Diphosphate
-
 Samoudi, Mojtaba
Institute of Industrial and Environmental Biotechnology, National Institute of Genetic Engineering and Biotechnology (NIGEB), Tehran, Iran, Tel: +98 21 44787359, E-mail: m.samoudi@nigeb.ac.ir
Samoudi, Mojtaba
Institute of Industrial and Environmental Biotechnology, National Institute of Genetic Engineering and Biotechnology (NIGEB), Tehran, Iran, Tel: +98 21 44787359, E-mail: m.samoudi@nigeb.ac.ir
-
Institute of Industrial and Environmental Biotechnology, National Institute of Genetic Engineering and Biotechnology (NIGEB), Tehran, Iran
-
Shahbani Zahiri, Hossein
-
Institute of Industrial and Environmental Biotechnology, National Institute of Genetic Engineering and Biotechnology (NIGEB), Tehran, Iran
-
Shariati, Parvin
-
Institute of Industrial and Environmental Biotechnology, National Institute of Genetic Engineering and Biotechnology (NIGEB), Tehran, Iran
Abstract: Background: Coenzyme Q10 (CoQ10) is an isoprenoid component used widely in nutraceutical industries. Farnesyl diphosphate synthase (FPPS) is a responsible enzyme for biosynthesis of farnesyl diphosphate (FPP), a key precursor for CoQs production. This research involved investigating the effect of FPPS over-expression on CoQs production in engineered CoQ10-producing Escherichia coli (E. coli).
Methods: Two CoQ10-producing strains, as referred to E. coli Ba and E. coli Br, were transformed by the encoding gene for FPPS (ispA) under the control of either the trc or PBAD promoters.
Results: Over-expression of ispA under the control of PBAD promoter led to a relative increase in CoQ10 production only in recombinant E. coli Br although induction by arabinose resulted in partial reduction of CoQ10 production in both recombinant E. coli Ba and E. coli Br strains. Over-expression of ispA under the control of stronger trc promoter, however, led to a severe decrease in CoQ10 production in both recombinant E. coli Ba and E. coli Br strains, as reflected by reductions from 629±40 to 30±13 and 564±28 to 80±14 µg/g Dried Cell Weight (DCW), respectively. The results showed high level of FPP reduces endogenous CoQ8 production as well and that CoQs are produced in a complimentary manner, as the increase in production of one decreases the production of the other.
Conclusion: The reduction in CoQ10 production can be a result of Dds inhibition by high FPP concentration. Therefore, more effort is needed to verify the role of intermediate metabolite concentration and to optimize production of CoQ10.
Introduction :
Ubiquinone, also known as CoQ, is an isoprenoid component and lipid-soluble molecule which plays various roles in both eukaryotic and prokaryotic cells. The most important function of CoQ is in energy generation, where it is responsible for electron transfer in the respiratory chain. In addition, CoQ has also been found to have an antioxidant function and is involved in various biological processes including gene expression, formation of disulfide bonds in proteins and generation of cellular signals 1-7. The lack of CoQ has been associated with various diseases 8.
Ubiquinone has been used as a medicine, but is now widely used as a nutritional supplement 9,10. The effect of ubiquinone on the lifespan of humans is currently being studied extensively while studies on other roles of ubiquinone are also underway 11. Ubiquinone is generally composed of a benzene ring and an isoprenoid side chain, which is a homopolymer of Isopentenyl diphosphate (IPP), also known as the isoprene unit. The length of the side chain determines the type of CoQ in various organisms. For example, the CoQ side chain in human cells is comprised of ten isoprene units and forms CoQ10, while Escherichia coli (E. coli) contains CoQ8 which possesses eight isoprene units in its side chain 12-16. The synthesis of the CoQ10 side chain is catalyzed by Dds, whereas Octaprenyl diphosphate synthase (Ods) is the enzyme responsible for biosynthesis of the corresponding side chain in E. coli. Therefore, the expression of the heterologous gene encoding for Dds can lead to the production of CoQ10 in transformed E. coli cells along with CoQ8. Accordingly, several CoQ10-producing E. coli strains have been devolved by introducing Dds encoding gene from various sources either via plasmid-expression system or chromosomal integration 4,15-18. The initial step in biosynthesis of isoprenoid side chain involves a condensation reaction between an IPP and its allelic isomer, Dimethylallyl diphosphate (DMADP) by Geranyl diphosphate synthase (GPPS) which results in the formation of Geranyl diphosphate (GPP) with ten carbons. FPPS adds another IPP to GPP and produces Farnesyl diphosphate (FPP) with 15 carbons. Likewise, Geranylgeranyl diphosphate synthase (GGPPS) uses FPP as a substrate and adds another IPP to form Geranylgeranyl diphosphate (GGPP) with 20 carbons. Finally, the polyprenyl diphosphate synthases such as Dds can use the above mentioned molecules (IPP, GPP, FPP and GGPP) as substrates for addition of more IPP molecules to make corresponding isoprenoid side chain. Finally, attachment of the synthesized side chain to benzene ring and subsequent modifications of the ring result in CoQ production 3,16,17,19,20. Figure 1 illustrates the biosynthetic steps for CoQ10 production.
Several Dds have been characterized and isolated from different sources 7,21-23. FPP is found to be the most preferred substrate for all types of Dds in reactions taking place in vitro 4. Accordingly, FPP can be an attractive precursor to control CoQ10 biosynthesis in engineered CoQ10-producing E. coli strains. Indeed, this suggests a hypothesis that the over-expression of the heterologous gene encoding for FPPS (ispA) under the control of various promoters in CoQ10-producing E. coli strains can control the biosynthesis of CoQ10. This strategy allows whole biosynthetic pathway productivity to be controlled through the intermediate metabolites levels.
The current study was conducted to determine the effect of high FPP concentration on both CoQ10 and CoQ8 accumulation in engineered E. coli cells. Two CoQ10-producing strains previously constructed by introducing Dds encoding gene from two different organisms 24 were used for concomitant expression of FPPS using either the trc or PBAD promoter to control the level of expression.
Materials and Methods :
Strain, media and culture condition: E. coli DH5α was selected as the host for construction of recombinant strains (Table 1). Luria-Bertani (LB) medium was used for cloning related cultures 25. For CoQ10 production, cells were initially grown in 5 ml of 2YT pre-culture medium (1% yeast extract, 1.6% tryptone and 0.5% NaCl) at 37°C. For production phase, seven ml of 2YTG medium (2YT medium supplemented with 0.5% glycerol and 0.01% 4-hydroxybenzoic acid) was used to carry out the cultivation in 25 ml tubes. The 2YTG media were inoculated with the pre-culture to an initial OD600 of 0.1, and were then incubated at 30°C for 48 hr in a rotary shaking incubator (GFL, UK) at 200 rpm. Ampicillin (100 μg/ml) and kanamycin (30 μg/ml) were added to the culture media as required. The growth of cells was then monitored by measuring optical density of the culture samples at 600 nm. The dry cell weight (g DCW/l) of the culture samples was calculated from a standard curve. CoQs quantification was determined in triplicate culture of recombinant strains.
Plasmid construction and transformation: All restriction enzymes were purchased from Fermentas (Germany). T4DNA ligase was purchased from New England Biolab (UK). Plasmid isolation and PCR product purification kits were from QIAGEN (US). The Dds encoding gene (dds) had previously been isolated from Agrobacterium tumefaciens (A. tumefaciens) (ATCC 33970) and Rhodobacter sphaeroide (R. sphaeroide) 2.4.1 (ATCC 17023), as reported elsewhere 23. The construction of plasmids harboring dds gene either from A. tumefaciens (atdds) or R. sphaeroide (rsdds) were described previously 24. Briefly, atdds was obtained from the existing pTatdds plasmid and amplified by PCR using Atdds-3 and Atdds-4 primers (Table 1). The PCR product was then double digested by XmaI and ligated into the pBBr1MCS2 plasmid which had been digested by the same restriction enzyme, resulting in the formation of the pBatdds plasmid. Likewise, rsdds gene was amplified by PCR using existing pTrsdds plasmid as template and Rsdds-F and Rsdds-R primers (Table 1). Next, the PCR product was double digested with EcoRI and Sac I and inserted into the pBr1MCS2 plasmid which had previously been digested with the same enzymes, leading to the formation of pBrsdds. FPPS encoding gene (ispA) had been isolated from the E. coli (GenBank: D00694.1) as reported previously 26. The ispA gene was obtained from the existing pBatdds ispA ubi CABG plasmid by PCR using ispA-F and ispA-R primers (Table1). The PCR product was subsequently double digested by EcoRI and BamHI and ligated to pTrc99A plasmid under the control of the trc promoter, leading to the formation of pTispA plasmid. The double digested ispA fragment was also ligated to pBAD24 plasmids under the control of PBAD promoter, using the same enzymes, and led to the formation of pDispA plasmid. Both pTrc99A and pBAD24 plasmids had previously been digested by EcoRI and BamHI. The recombinant plasmids were introduced to E. coli cells using a modified chemical transformation method 27.
CoQs extraction: CoQs content of cells was determined by extraction. Specifically, 500 μl of culture was centrifuged at 10,000 xg (Heraeus, Biofuge, UK). The pellet was washed with 1 ml of distilled water, then 1 ml of 20 mM Tris-HCl buffer (pH=7.6). 450 µl of lysis buffer [8% sucrose, 5% Triton X-100, 50 mM Tris-HCl (pH=8), 50 mM EDTA (pH=8) and 1 mg/ml of lysozyme] was added to washed pellet and incubated at 37°C for 30 min. 900 μl of a hexane/propanol (5:3 v/v) mixture was added to lysed cells and centrifuged at 11,000 xg for 2 min. The hexane organic phase containing CoQs was transferred to a clean tube. Hexane (500 µl) was added to the aqueous phase and centrifuged at 11,000 xg for 2 min. The second hexane phase was added to the first and dried in a vacuum evaporator (Speedvac®, AES 1010, US). The pellet was dissolved in 500 μl of absolute ethanol (HPLC grade, Merck, US).
Determination and quantification of CoQs: The CoQ content was determined from 10 μl injected sample by HPLC (Shimadzu 10A system, Japan) equipped with a Symmetry® C18 column (Waters, US). The mobile phase was the mixture of ethanol and methanol (70:30 v/v) with flow rate of 1 ml/min. CoQs were detected using a UV detector at 275 nm. The identification of corresponding peaks in the HPLC chromatograms of experimental samples was carried out using authentic standards, CoQ9 and CoQ10. Quantification of CoQs were performed with the aid of an appropriate standard curve.
Results :
CoQs production in E. coli: As mentioned in our previous work 24, two type of dds genes, rsdds and atdds, were inserted into the pBr1MCS2 plasmid, and resulted in the formation of plasmids pBrsdds and pBatdds. The recent plasmids were transformed into E. coli DH5α, and designated as E. coli Br and E. coli Ba, respectively. The recombinant strains were grown in 2YTGH medium under the same experimental conditions. Their CoQs content was extracted and determined quantitatively by detection of the corresponding peaks in the HPLC chromatogram. The expression of rsdds or atdds in the transformed E. coli Ba and E. coli Br led to the production of CoQ10 together with CoQ8. On average, E. coli Ba had CoQ10 content of 629±40 μg/g DCW while E. coli Br had 564±28 μg/g DCW under the described experimental conditions. E. coli naturally has an Ods encoding gene. So, the CoQ8 content was also measured in CoQ10-producing strains. Expression of dds led to a significant decrease in CoQ8 production by both E. coli Ba (from 990±32 to 360±23 μg/g DCW) and E. coli Br (from 990±32 to 246±24 μg/g DCW), when compared to that in E. coli DH5α 24. An example of HPLC chromatogram for CQs identification in E. coli Ba is shown in figure 2. The recombinant E. coli strain produced CoQ10 and slight amounts of CoQ9 in addition to naturally occurring CoQ8. The CoQ9 might be produced as a result of immature release of a fraction of the growing polyprenyl chains from the Dds 17.
Effect of ispA over-expression under the control of PBAD promoter in CoQ10-producing E. coli: The expression of ispA results in an increased level of FPP production. FPP is the most suitable substrate for all types of Dds in vitro reactions 4. In order to study the effect of FPPS on CoQs production, the ispA gene was ligated into the pBAD24 plasmid, leading to the formation of pDispA plasmid (Figure 3A). This recombinant plasmid was used for transformation of E. coli Ba and E. coli Br. The resulting recombinant cells, designated as E. coli BaDi and E. coli BrDi, were cultured in 2YTG medium, and their CoQs were extracted and quantified by HPLC.
In the absence of inducer, the expression of ispA under the control of PBAD promoter does not have significant effect on CoQ10 production in E. coli BaDi, with negligible change from 629±40 to 636±35 μg/g DCW (Figure 4), while relative increase was observed in E. coli BrDi from 564±28 to 750±28 μg/g DCW (Figure 5). However, once induction by arabinose was applied, relative reduction in CoQ10 production was obtained which was proportional to the concentration of arabinose in both strains. In the absence of induction, the CoQ8 production demonstrated roughly a twofold reduction in E. coli BrDi (from 246±24 to 117±13 μg/g DCW), while no significant change (from 361±23 to 373±28 μg/g DCW) was observed in E. coli BaDi. Insignificant reduction in CoQ8 production was observed in both strains with increased arabinose concentration (Figures 4 and 5). As a whole, the results showed that CoQs production in both ispA-expressing strains is inhibited once the induction is applied.
Effect of ispA over-expression under the control of trc promoter in CoQ10-producing E.coli: In order to confirm that the reduction in CoQs production is due to the inhibitory effect of high FPP concentration, ispA was expressed under the control of trc promoter which is a stronger promoter than PBAD and regulated alternatively by adding IPTG to the culture media. So, the ispA gene was ligated into the pTrc99A plasmid, leading to the formation of pTispA (Figure 3B). This recombinant plasmid was used for transformation of E. coli Ba and E. coli Br. The resulting recombinant cells, designated as E. coli BaTi and E. coli BrTi, were cultured in 2YTG medium under the same conditions, as described previously. Then, their CoQs were extracted and quantified by HPLC. The results of this study demonstrated that the over-expression of ispA was highly inhibiting for CoQ10 biosynthesis in both E. coli BaTi (Figure 6) and E. coli BrTi (Figure 7) and reduced CoQ10 production levels in the two strains from 629±40 to 289±16 μg/g DCW and from 564±28 to 110±20 μg/g DCW, respectively, even in the absence of an inducer. The presence of the inducer, IPTG (0.05 mM), led to a further decrease in CoQ10 production to 30±13 μg/g DCW in E. coli BaTi and 80±14 μg/g DCW in E. coli BrTi. These results show that the inhibition of CoQ10 production was more severe when compared to the E. coli BrDi and E. coli BaDi where the ispA gene is controlled by PBAD promoter.
In addition, it was observed that the reduction of CoQ10 production in both E. coli BaTi and E. coli BrTi occurred simultaneously with an increase in CoQ8 production when compared to E. coli Ba and E. coli Br (from 360 to 890 μg/g DCW in E. coli BaTi and 246 to 580 μg/g DCW in E. coli BrTi). However, induction by IPTG (0.05 mM) decreased the CoQ8 biosynthesis as well in both E. coli BaTi (to 750 μg/g DCW) and E. coli BrTi (to 310 μg/g DCW) as shown in figures 6 and 7. The growth of recombinant strains was also studied and showed no significance changes by ispA over-expression (Figure 8).
Discussion :
Previous isoprenoid research has implicated the critical role of IPP levels in the biosynthesis of isoprenoid including CoQs 28-31. Accordingly, various strategies have been used to improve CoQ10 production through IPP availability by introducing genes that encode for mevalonate biosynthetic pathway enzymes 28,32-35. Although these efforts have led to an increase in CoQ10 accumulation, there has not yet been an effective method developed 17. Specifically, it has been demonstrated that other intermediate metabolites alongside IPP contribute to the control of ubiquinone biosynthesis 36.
IPP, GPP, FPP and GGPP are potential substrates for the stepwise addition of IPP by Dds to make the long 50-carbon chain of CoQ10. Previous researches have indicated that FPP is the most suitable substrate for the Dds reactions in vitro 4. In this study, FPPS encoding gene was introduced into two CoQ10-producing strains under the control of either the trc or PBAD promoters. The ispA gene was expressed under the control of PBAD promoter by which expression can be modulated over a wide range of inducer (arabinose) concentrations. The results showed relative increase in CoQ10 production in E. coli BrDi when no induction was applied. Induction by arabinose, which results in higher FPP level, led to reduction in CoQ10 accumulation in both strains. Thus, the higher FPP concentrations may have negative impact on CoQ10 accumulation. The over-expression of ispA caused a large decrease in CoQ10 production when the trc promoter was employed (E. coli BaTi and E. coli BrTi). In fact, the higher FPP concentrations from high-copy plasmid with strong trc promoter led to more reduction in CoQ10 production, confirming the hypothesis that high concentration of FPP inhibits CoQ10 accumulation; probably through inhibition of Dds enzymatic activity.
When it comes to monitor the simultaneous CoQ8 production, it was observed that the production of CoQ10 brings about a decline in CoQ8 production and vice versa. It can be assumed that the introducing of dds gene leads the substrate flow to be conducted towards CoQ10 production, thus decreasing CoQ8 biosynthesis in E. coli Ba and E. coli Br. In contrast, due to the negative effects of ispA gene expression on CoQ10 production in ispA-producing strains, the substrate flow might be conducted again towards CoQ8 biosynthesis to some extent, because FPP can also function as a substrate for Ods as well as Dds 4. However, higher level of FPP derived from induction by IPTG or higher level of arabinose reduced CoQ8 production as well as that of CoQ10 accumulation. It seems that high concentrations of FPP may have an inhibitory effect on the E. coli Ods enzyme activity as well as Dds, thus causing a substantial decrease in CoQ8 biosynthesis. Growth was not significantly affected by the expression of ispA. It seems that the residual amount of CoQ8 produced in ispA-expressing strains is still sufficient for respiration and consequently supporting the growth (Figure 8).
It seems that biochemical properties of various Dds from different organisms also play role in determining their enzymatic activities. For example, introduction of pDispA in E. coli BrDi resulted in relative increase in CoQ10 production (from 546 to 750 μg/g DCW) while it had no significant effect on CoQ10 production in E. coli BaDi (from 629 to 636 μg/g DCW). This may have resulted from the relatively low similarity between the enzymes, which is reflected in differences in their biochemical characteristics 23.
It seems that the biosynthesis of CoQs is controlled by intermediate metabolite concentration in a complex way that inhibition of polyprenyl diphosphate synthases by precursors is a major bottleneck and adjusting the precursor’s concentrations is of great importance. Different strategies can be employed to achieve desirable precursor's concentration in engineered E. coli. For example, it is proposed that the type of expression vectors and the promoter strength are very important for CoQ10 production in recombinant E. coli 6. This idea was confirmed in the current study where pTrc99A plasmid, a high-copy number vector with strong promoter, was found to dramatically reduce CoQ10 production while using pBAD24 plasmid with an inducible weaker promoter has no severe, reducing effect on CoQ10 production and even relatively increased CoQ10 production in E. coli BrDi. It has also been observed that the co-production of other isoprenoid compounds which share same intermediate precursors may also contribute to control biosynthesis of CoQs through controlling metabolite flow rate 24. These findings suggest that the over-expression of the dds gene with a high-copy plasmid, which leads to higher Dds concentration, alongside the ispA may enhance the CQ10 production via quenching the inhibition caused by the high concentration level of FPP.
Conclusion :
In summary, the regulation of the metabolite flow rate is a crucial step, and thus highly important from a biotechnological perspective. Therefore, more effort is needed to verify the role of intermediate metabolite concentration in the biotechnology strategy and to optimize the methods through which the production of CoQs can be elevated.
Acknowledgement :
We hereby thank Dr. Fatemeh Tabandeh and Dr. Sarah W. Harcum for their scientific editing of the work. This work was supported by National Institute of Genetic Engineering and Biotechnology (NIGEB).
Conflict of Interest :
None of the authors has any conflicts of interest to declare.
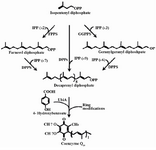
Figure 1. The tandem condensation reactions by decaprenyl diphos-phate synthase result in the polymerization of isopentenyl diphos-phate (IPP) molecules into decaprenyl diphosphate. The indicated enzymes are as follow: FPPS, farnesyl diphosphate synthase; GGPPS, geranylgeranyl diphosphate synthase; DPPS, decaprenyl diphosphate synthase and UbiA, 4HB-polyprenyl transferase (Zahiri et al, 2009).
|
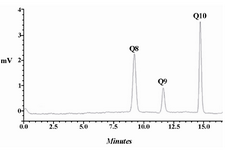
Figure 2. HPLC chromatogram for CQs identification in E. coli Ba. The recombinant E. coli produced CoQ10 and slight amounts of CoQ9 in addition to naturally occurring CoQ8.
|
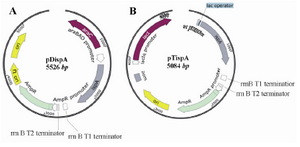
Figure 3. Schematic representation of recombinant pDispA plasmid, encoding for ispA gene under the control of PBAD promoter (A) and pTispA plasmid, encoding for ispA gene under the control of trc promoter B). The genetic maps are generated by SnapGene software (from GSL Biotech; available at snapgene.com).
|
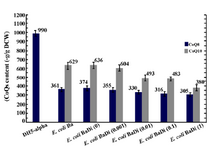
Figure 4. Plasmid pDispA was introduced into E. coli Ba, and the coenzyme Qs content was quantified in the resulting strain, as referred to E. coli BaDi. Numbers in brackets indicate arabinose mM concentration in cultures. Error bars indicate the standard error of the mean of three independent experiments.
|
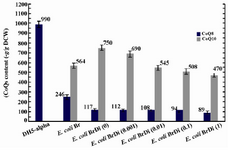
Figure 5. Plasmid pDispA was introduced into E. coli Br, and the coenzyme Qs content was quantified in the resulting strain, as referred to E. coli BrDi. Numbers in brackets indicate arabinose mM concentration in cultures. Error bars indicate the standard error of the mean of three independent experiments.
|
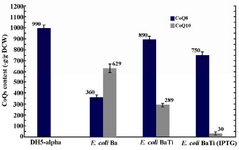
Figure 6. Plasmid pTispA was introduced into E. coli Ba, and the coenzyme Qs content was quantified in the resulting strain, as referred to E. coli BaTi. Error bars indicate the standard error of the mean of three independent experiments.
|
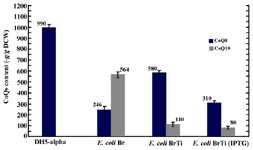
Figure 7. Plasmid pTispA was introduced into E. coli Br, and the coenzyme Qs content was quantified in the resulting strain, as referred to E. coli BrTi. Error bars indicate the standard error of the mean of three independent experiments.
|
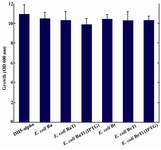
Figure 8. No significant changes were observed in growth associated with ispA over-expression. Error bars indicate the standard error of the mean of three independent experiments.
|

Table 1. Strains, plasmids and primers used in this study
|
|