Inactivation of aprE Gene in Bacillus subtilis 168 by Homologus Recombination
-
 Rabbani, Mohammad
Department of Pharmaceutical Biotechnology, School of Pharmacy and Pharmaceutical Sciences, Isfahan Pharmaceutical Sciences Research Center, Isfahan University of Medical Sciences, Isfahan, Iran, Tel: +98 311 7922646; Email: rabanim@yahoo.com
Rabbani, Mohammad
Department of Pharmaceutical Biotechnology, School of Pharmacy and Pharmaceutical Sciences, Isfahan Pharmaceutical Sciences Research Center, Isfahan University of Medical Sciences, Isfahan, Iran, Tel: +98 311 7922646; Email: rabanim@yahoo.com
-
Department of Pharmaceutical Biotechnology, School of Pharmacy and Pharmaceutical Sciences, Isfahan Pharmaceutical Sciences Research Center, Isfahan University of Medical Sciences, Isfahan, Iran, Tehran, Iran
-
Soleymani, Safoura
-
Department of Pharmaceutical Biotechnology, School of Pharmacy and Pharmaceutical Sciences, Isfahan Pharmaceutical Sciences Research Center, Isfahan University of Medical Sciences, Isfahan, Iran, Tehran, Iran
-
Mir Mohammad Sadeghi, Hamid
-
Department of Pharmaceutical Biotechnology, School of Pharmacy and Pharmaceutical Sciences, Isfahan Pharmaceutical Sciences Research Center, Isfahan University of Medical Sciences, Isfahan, Iran, Tehran, Iran
-
Soleimani, Narjes
-
Department of Pharmaceutical Biotechnology, School of Pharmacy and Pharmaceutical Sciences, Isfahan Pharmaceutical Sciences Research Center, Isfahan University of Medical Sciences, Isfahan, Iran
-
Moazen, Fatemeh
-
Department of Pharmaceutical Biotechnology, School of Pharmacy and Pharmaceutical Sciences, Isfahan Pharmaceutical Sciences Research Center, Isfahan University of Medical Sciences, Isfahan, Iran, Tehran, Iran
Abstract: Background: One of the most important producers of high quality industrial enzymes is the Gram-positive bacterium, Bacillus subtilis (B. Subtilis). One major limitation that hinders the wide application of B. subtilis is the secretion of high levels of extracellular proteases which degrade the secreted foreign proteins. In this study, homologus recombination technique was used to knock out its protease gene, aprE.
Methods: The internal segment of the pro-sequence of aprE gene of B. subtilis 168 with a length of 80 bps and its complementary sequence were synthesized and ligated into pUB110 at EcoR1 and XbaI restriction sites. Competent cells of B. subtilis 168 were prepared and transformed by electroporation using Bio Rad gene pulser as explained in the methods section. Transformants carrying the recombinant plasmid were selected for resistance to neomycin. The success of homologous recombination was checked by PCR amplification of the neomycin gene which was part of the vector and did not exist in the genome of B. subtilis 168. The protease activity was measured using the Protease Fluorescent Detection Kit based on the proteolytic hydrolysis of fluorescein isothiocyanate (FITC)–labeled casein-substrate.
Results: The results demonstrated that aprE gene would not be able to produce further active subtilisin E. The reduction of protease activity also confirmed the efficacy of the induced mutation in this gene.
Conclusion: It will therefore be a major challenge for future research to identify and modulate quality control systems of B. subtilis which limit the production of high quality protease- sensitive products such as lipase.
Introduction :
Bacillus subtilis is a well-known Gram-positive soil bacterium that naturally produces and secrets high concentrations of proteins into the medium 1. The absence of an outer membrane in B. subtilis can simplify the protein secretion pathways and allow the organism to secrete high levels of extracellular proteins. In contrast to the Gram-negative bacterium Escherichia coli, B. subtilis is considered as a GRAS organism (Generally Recognized As Safe). Thus, scientists have widely used B. subtilis for commercial exploitation as a major "cell factory" for the secretion of heterologous proteins 2,3. As far as the capability of secreting extracellular enzymes directly into the culture medium is concerned, B. subtilis can potentially serve as an efficient expression host 4,5. The secreted foreign proteins usually remain in biologically active forms, and the downstream purification is greatly simplified 6,7.
One of the major limitations that hinder the wide application of B. subtilis is the secretion of high levels of extracellular proteases which degrade the secreted foreign proteins. It is well established that B. subtilis has six extracellular proteases including neutral protease A, subtilisin (also known as alkaline protease), extracellular protease, metalloprotease, bacillopeptidase F, and neutral protease B 8. In view of the fact that the protease production is limited, protease deficient B. subtilis strains have been developed by genome engineering techniques 9. For example, B. subtilis WB800, deficient in eight extracellular proteases can serve as an excellent host for the expression of heterologous proteins 10.
In order to improve the production of heterologous proteins in B. subtilis 168, it was decided to inactivate the aprE gene of this bacterium encoding one of the major extracellular alkaline serine proteases, Subtilisin E, by site directed mutation of this gene using homologous recombination techniques. For this purpose, the pro-sequence in the aprE gene was targeted. Subtilisin E is first made as pre-pro-subtilisin 9,11,12. This enzyme consists of a signal peptide for protein secretion (pre-sequence) and a peptide extension of 77 amino acid residues (pro-sequence) located between the signal peptide and the mature protease 13. Studies have indicated that the pro-sequence is essential for guiding appropriate folding of the enzymatically active conformation of Subtilisin E 12,13. Inactivating the pro-sequence of the aprE gene, therefore, should impair the production of an active subtilisin resulting in a genetically engineered strain deficient in one of the major extracellular proteases making an appropriate host for the expression of protease sensitive products.
Materials and Methods :
Bacterial strains, plasmids, and growth conditions: The bacterial strain used in this study was B. subtilis strain 168 (DSMZ, Germany) containing the plasmid pUB110. The medium consisted of 7% maltose (w/v), 0.05% yeast extract (Difco), 4.6% dried corn steep liquor, 0.5% fish meal extract, 0.1% KH2PO4, 0.02% MgSO4• 7H2O, and 15 μg/ml tetracycline (pH=6.8). The supernatant was collected from the culture broth by centrifugation at 10,000 g for 20 min and after that the pellet was resuspended in 1 mM EDTA, 1 mM PMSF, 1 mM DTT, and 5% glycerol in 50 mM potassium phosphate buffer (pH=7.4). Next, it was sonicated (model 7500; BIOMIC). The supernatants were then collected following centrifugation. Plasmid DNA from B. subtilis 168 was prepared as described by Sambrook et al 14.
Insert preparation: The internal segment of the pro-sequence of the aprE gene of B. subtilis 168 with a length of 80 bps (nucleotides 96 to 176 upstream from the initiation codon) and its complementary sequence were synthesized by Kawsar Biotech Company (Iran). Restriction sites for the enzymes EcoRI and XbaI were designed on both ends of the single strands as outlined below:
5´AATTCagcagtacagaaaagaaatacattgtcgtcggatttaaacagacaatgagtgccatgagttccgccaagaaaaaggatgtT3´ and 5´CTAGAacatcctttttcttggcggaactcatggcactcat tgtctgtttaaatccgacaatgtatttcttttctgtactgctG3´
Vector preparation: Twenty five µl of PUB110 was cross digested with the enzymes EcoRI and XbaI in a total volume of 100 µl according to the manufacturer’s recommended conditions (Germany). Gel electrophoresis was also run to verify complete digestion of the plasmid PUB110.
Ligation reaction and transformation: The ligation reaction was carried out using an appropriate amount of vector and insert (a ratio of 3:1 insert to vector). One µl of 10ligation buffer and 1 µl of Ligase were added to the mixture adjusting the final volume to 10 µl with ddH2O. The ligation mixture was incubated overnight at 16C.
Competent cells of B. subtilis 168 were prepared and transformed by electroporation using Bio Rad gene pulser. Electrocompetent cells were prepared by diluting 2 ml of the overnight culture of B. subtilis 168 in 50 ml of fresh LB culture medium and grown at 37C to reach OD value of 0.3 at 600 nm. Then the cell suspension was cooled on ice for 10 min and harvested by centrifugation at 4C and 10000 g for 10 min. The cells were suspended in 5 ml ice-cold electroporation buffer and spun at 4C at 10000 g for 10 min. Finally, the cells were suspended in 1.5 ml of ice-cold electroporation solution and the competent cells were kept on ice until use. Ten µl (5 ng/µl) of the ligation mixture containing the recombinant plasmid pUB110 was added to 100 µl of electrocompetent cells and homogenized by gently mixing with pipette several times. The mixture was then transferred into a pre-chilled cuvette and an electric shock with a voltage of 1000 V and a time constant of 8.6 ms was applied. Following the electric shock, 2 ml of LB culture medium was immediately added to the electroporated cells followed by incubation for 1.5 hr at 37C.
Selection for neomycin resistance: Transformants carrying the recombinant plasmid were selected for resistance to neomycin.
Primer design for the neomycin gene and PCR amplification: To confirm the incidence of homologous recombination, PCR amplification of the neomycin gene was carried out. Neomycin gene was part of the plasmid pUB110 used as the vector and it did not exist in the genome of B. subtilis 168. The neomycin gene was amplified using the genomic DNA from transformed B. subtilis 168 as a template with a pair of primers, F (5΄-TCGGAAAGTTGA CCAGA-3΄) and R (5΄-TTTGTGCCCTTATC GTAG-3΄). PCR was conducted by subjecting the reaction mixture to an initial denaturation at 94C for 5 min, followed by 30 cycles of 94C for 1 min, 55C for 2 min, 72C for 3 min and a final extension step at 72C for 5 min.
Extracellular protease activity assay: To quantitatively compare the protease activity of wild and mutant strains deficient in active subtilisin E, the Protease Fluorescent Detection Kit (Sigma-Aldrich, Germany) based on the proteolytic hydrolysis of a fluorescein isothiocyanate (FITC)-labeled casein-substrate was used according to the manufacturer’s protocol. FITC-casein is native casein that has been labeled using fluorescein isothiocyanate. The UV absorbance of this heavily-labeled, intact protein substrate at 280 nm changed dramatically upon digestion by proteases, resulting in a measurable indication of proteolysis. Different concentrations of FITC-casein were also prepared by serial dilutions and their absorbance at 280 nm was measured to determine the concentration ranges across which the Beer-Lambert law was obeyed.
Results :
Construction of recombinant plasmid, pUB110: To prepare the pUB110for ligation, the plasmid was first subjected to restriction digestion with the enzymes EcoRI and XbaI. Digested plasmid was then dephosphorylated by alkaline phosphatase and recovered from the gel. The internal segment of pro-sequence in the aprE gene of B. subtilis was constructed with the same restriction enzyme sites (EcoRI and XbaI) as the vector. Agarose gel analysis of the pUB110digested plasmid after dephosphorylation and EcoRI and XbaI digested insert is depicted in figure 1. Constructed insert and vector had the lengths of 80 and 4000 bps, respectively. An appropriate amount of prepared plasmid (pUB110) and insert were mixed for ligation reaction with T4 DNA ligase. Then recombinant plasmid was transformed into B. subtilis 168. Figure 1 shows the pUB110plasmid digested with EcoRI and XbaI and also the 80 bp insert. The integrity of the plasmid (4 kb) of cut plasmid and the purchased insert both were verified.
Transformation of recombinant pUB110plasmid into Bacillus subtilis 168: Screening the electroporated cells was done for antibiotic resistance and performed on nutrient agar plates supplemented with 5 and 10 mg/ml of neomycin 14. The colonies of transformed cells from the plasmid containing neomycin resistance gene successfully grew and formed a bacterial lawn on LB media as shown in figure 2. DNA extracted from the transformed cells was compared with non-transformed bacteria. Figure 3 shows the pattern of DNA extracted from the transformed and non-transformed bacteria.
PCR amplification of neomycin gene: To confirm the incidence of homologous recombination, PCR amplification of the neomycin gene was carried out. Neomycin gene was part of pUB110and it did not exist in the genome of B. subtilis 168. Integration of this plasmid in the chromosome by double cross-over replaced the neomycin resistant gene in chromosomal DNA of transformed B. subtilis 168. The neomycin gene was amplified using genomic DNA from transformed B. subtilis 168 as a template with a pair of designed primers. A sharp band in the 300 bps region (Figure 4) in the transformed cells proved the integration of the plasmid DNA into chromosomal DNA of transformed bacteria.
Extracellular protease activity assay in wild and mutant strains: Protease activity of samples was determined by measuring the residue of FITC-Casein in media. Activity measurement at the examined condition indicated that in mutant samples, the UV absorbance was increased in comparison with wild samples at 280 nm. These changes demonstrated that the aprE gene in B. subtilis 168 would not further be able to produce active subtilisin E and finally would decrease the protease activity.
Discussion :
B. subtilis bacteria utilize especial systems that secrete proteins out of cells. Subtilisin E is one of typical examples which functions similarly 15. This extracellular alkaline serine protease encoded with aprE gene was produced at the end of exponential growth phase 16. Subtilisin E leads to degradation of other important proteins like lipase enzymes. It is known that if aprE gene is knocked out, B. subtilis will no longer be able to produce active subtilisin E and therefore, it can help to produce active proteins which are sensitive to proteases 13.
The aim of the present study was the inactivation of aprE gene using site directed mutation at pro-sequence region. For this purpose, the recombinant plasmid was constructed with mixing appropriate amount of insert and pUB110 plasmid as a vector. After transformation of recombinant plasmid into B. subtilis 168, screening results showed that electroporated cells can grow in supplemented media containing neomycin antibiotic. Comparison between transformed and non-transformed cells demonstrated that digestion, ligation and transformation were done successfully. The results of PCR amplification of neomycin gene further confirmed the homologous recombination. Finally, in accordance with our finding, the reduction of protease activity demonstrated that the mutation in aprE gene has indeed taken place at the right site.
Conclusion :
As previously mentioned, B. subtilis is one of the most widely used bacterium for the production of industrial enzymes but its applications is limited due to its proteases. According to the results obtained in this study, homologous recombination techniques can help to inactivate protease encoding genes and improve production of other protease-sensitive- proteins. It will therefore be a major challenge for future research to identify and modulate quality control systems of B. subtilis which limit the production of high quality protease- sensitive products such as lipase.
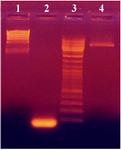
Figure 1. PCR amplification of 80 bp insert from digested plasmid pUB110 with EcoRI and XbaI. HindIII digested lambda DNA, the 80 bp insert, DNA ladder and pUB110plasmid digested with EcoRI and XbaI are shown in lanes 1, 2, 3 and 4, respectively
|
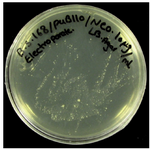
Fiagure 2. The colonies of transformed cells from the plasmid containing neomycin resistance gene streaked out on LB plates
|
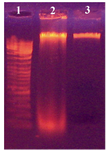
Figure 3. Electrophoretic analysis of DNA from transformed and non-transformed B. subtilis 168. Lanes; 1) DNA size marker; 2) Non-transformed cells and 3) Transformed cells
|
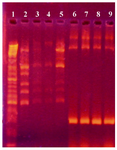
Figure 4. Agarose gel electrophoresis showing PCR amplification products of neomycin gene (300 bp). First lane is a DNA size marker (100 bp DNA ladder). Lanes 2-5 and lanes 6-9 are PCR products from the neomycin gene in non-transformed and transformed cells, respectively
|
|