Development of Polymer-coated Glass Slides as Optical Oligonucleotide Microarrays
-
Pourjahed, Atefeh
-
Biomaterials Group, Faculty of Biomedical Engineering, Amirkabir University of Technology, Tehran, Iran
-
 Rabiee, Mohammad
Biomaterials Group, Faculty of Biomedical Engineering, Amirkabir University of Technology, Tehran, Iran, Tel: +98 21 64542381; E-mail:mrabiee@aut.ac.ir
Rabiee, Mohammad
Biomaterials Group, Faculty of Biomedical Engineering, Amirkabir University of Technology, Tehran, Iran, Tel: +98 21 64542381; E-mail:mrabiee@aut.ac.ir
-
Biomaterials Group, Faculty of Biomedical Engineering, Amirkabir University of Technology, Tehran, Iran
-
Tahriri, Mohammadreza
-
Biomaterials Group, Faculty of Biomedical Engineering, Amirkabir University of Technology, Tehran, Iran
Abstract: Background: The microarray technology is in needed of cost-effective, low background noise and stable substrates for successful hybridization and analysis.
Methods: In this research, we developed a three-dimentional stable and mechanically reliable microarray substrates by coating of two polymeric layers on standard microscope glass slides. For fabrication of these substrates, a thin film of oxidized agarose was prepared on the Poly-L-Lysine (PLL) coated glass slides. Unmodified oligonucleotide probes were spotted and immobilized on these double layered thin films by adsorption on the porous structure of the agarose film. Some of the aldehyde groups of the activated agarose linked covalently to PLL amine groups; on the other side, they bound to amino groups of adsorbed tail of biomolecules. These linkages were fixed by UV irradiation at 254 nm using a CL-1000 UV. These prepared substrates were compared to only agarose-coated and PLL-coated slides.
Results: Atomic Force Microscope (AFM) results demonstrated that agarose provided three-dimensional surface which had higher loading and bindig capacity for biomolecules than PLL-coated surface which had two-dimensional surface. The nano-indentation tests demonstrated the prepared double coating was more reliable and flexible for mechanical robotic spotting. In addition, the repeated indentation on different substrates showed uniformity of coatings. The stability of novel coating was sufficient for hybridization process. The signal-to-noise ratio in hybridization reactions performed on the agarose-PLL coated substrates increased two fold and four fold compared to agarose and PLL coated substrates, respectively.
Conclusion: Finally, the agarose-PLL microarrays had the highest signal (2920) and lowest background signal (205) in hybridization, suggesting that the prepared slides are suitable in analyzing wide concentration range of analytes.
Introduction :
Microarray technology is a potential tool in biomolecular research, which provides high sensitivity and specificity in genomic and proteomic tests 1-4. One of the important challenges in applying this technology is the surface chemistry of substrate. The chemistry should be active and compatible for spotting and immobilization of different probe biomolecules (DNA, proteins, phospholipids and cells) 5. Glass, the most widely used microarray substrate, is inexpensive and inert and has low background signal under fluorescence detection 6,7. In addition, glass can be easily modified 8,9 to increase binding capacity, hybridization efficiency, spot uniformity and stability of immobilized probes 5. Various modification methods activate glass surface for attachment of biomolecules as diagnostic probes. Depending on the properties of the coating, physical or chemical interactions may occur between probe and modified surface of the glass. PLL and aminosilane are physically attached to glass substrate and do not require probe modification 9,10. However, these substrates have low binding capacity and flat surfaces which result in sterical limitations in hybridization of the target to probes 10. To solve this problem, we can elevate the probes from flat surface; this can increase binding capacity of the glass 11,12. Polymers such as poly (acrylamide) or agarose can act as dendrimetric spacers which can elevate probes from substrate and show high hybridization efficiency 13-17.
Agarose is inexpensive, non-toxic 18,19 and commercially available. Coating of oxidized agarose on glass slides can provide aldehyde groups which bind to amino-modified DNA probes 19; also DNA probes can attach to agarose-coated glass surface using ultraviolet (UV) radiation 19,20.
In this work, we prepared PLL, agarose and PLL/agarose coatings on the standard glass slides as oligonucleotide microarray substrates. The probes were immobilized using UV radiation. The prepared slides were compared together and finally the obtained results ascertained the potential of the slides as DNA microarray substrate.
Related works: Dufva et al introduced standard protocol for grafting agarose layer on to unmodified glass microscope slides. AFM characterization of grafted agarose film demonstrated a significant increase in surface roughness. SEM analysis showed the porous structure of agarose. Amino-modified 21 and 25 bp long capture probes were immobilized on activated agarose surface. Agarose-coated slides had the highest signal-to-noise ratio in hybridization. In addition, UV immobilization of DNA probes on to agarose surface on glass slides further increased the signal-to-noise ratio 19.
In the research of Wei et al a thin film of oxidized agarose was prepared on the surface of ordinary glass slides as antibody/antigen microarray substrate. The aldehyde groups derived from the agarose film were covalently linked with amino groups of antigens. AFM characterization and assay results demonstrated that gel assembly film was a surface of three-dimensional structure. The enzyme immunoassay system on the substrates was successfully developed. These microchips could provide simultaneous analysis for multi-autoantibodies 21.
Koch and his co-workers spotted amine-modified oligonucleotides on the surface of activated agarose-coated slides using a manual eight-pin arraying device. The probes easily distinguished different plant pathogens from each other without cross section. Spotted probes on agarose differentiated samples as readily as probes on nylon but with potentially higher spot density and gave much higher signal than probes on silanated slides 22.
In another study, Dufva et al ascertained that 13- to 17- base oligonucleotide tagged with a poly(T) 10-poly(C) 10 tail, but otherwise unmodified, could be crosslinked by UV irradiation to an agarose film grafted on to unmodified glass. These microarrays were used to diagnose mutations in the human β-globin gene. Sufficiently high discrimination signals were obtained between perfect match and mismatch probes in hybridization.
Xu et al developed a new kind of sensitive substrates by coating agarose on silica opal film fabricated on glass substrate. These substrates provided stronger fluorescence signals compared with agarose on unmodified glass and thus improved detection sensitivity 23.
Materials and Methods :
Materials: Microscope glass slides (25.4 mm×76.2 mm×1 mm) and glass cover slips were purchased from Sigma-Aldrich. PLL was purchased from Sigma and Agarose was provided by Roche. Betaine, Sodium Diodecyl Sulfate (SDS), Succinic Anhydride (SA), N-Methyl 2-Pyrrolidone (NMP), Boric acid, Sodium hydroxide (NaOH), Ethanol, Bovine Serum Albumin (BSA), Formamide and Salmon sperm DNA were purchased from Sigma. Saline sodium citrate was provided by Ambion. Sodium Periodate was purchased from Fluka. DNA extraction kits were purchased from Tadbir Fan Azma Co. Cy5mono-reactive dye was purchased from GE-Healthcare. Finally, DNA labeling kits were provided by Agilent Technologies Co.
Preparation of polymer film-coated slides: Three different polymeric layers were coated on the standard glass slides: (i) PLL, (ii) Agarose, and (iii) Agarose layer on the PLL coating.
After cleaning of microscope slides by a mixed solution of NaOH and ethanol, the slides were rinsed in dH2O. For coating of PLL polymer on cleaned slides, PLL fresh coating solution was prepared by mixing 1:9 ratio of PLL and dH2O, respectively. The slides were covered with the solution for 1 hr on an orbital shaker (50 rpm, Cole Parmer Economic orbital shaker). The slides were transferred in to slide centrifuge (Arrat It® microarray high speed centrifuge) for liquid collection. Then, slides were placed in vaccum oven (43ºC, Memmert) for complete drying. Agarose layer was also coated on unmodified glass slides and PLL-coated glass slides according to method described by Dufva et al 19.
Preparation of microarrays: DNA probes, which have 35-40 bp long (Table 1) were spotted on the fabricated substrates using Q-array mini system (Genetix, Germany). Printing solution, containing 150 mM of phosphate buffer, pH=8.5 and 3X SSC with 1.5 M Betaine wasprepared. Dried probes were suspended in the mixture of equal volume of printing buffer and dH2O to obtain final concentration of ~ 200 ng/µl.
Substrates were printed at 40-50% relative humidity, and then dried in the air for 10 min. The probes were spotted in quadruplicate on the substrates. The DNA probes were immobilized on the slides by UV radiation at 254 nm using a CL-1000 UV crosslinker (UVP Inc.) for 3 min. Then, arrays were immersed in 1% Sodium Diodecylsulphate (SDS) for 30 s. Finally, the slides were dried using compressed nitrogen gas.
Morphological characterization of the coated slides: The micro-scale micrographs of the spotted and unspotted glass slides were obtained using AIS2100 Scanning Electron Microscopy (Seron Technology Inc., Korea). A Hysitron Inc. (USA) Triboscope® Nanomechanical test instrument with 2D transducer, complete software and Berkovich diamond indenter were used to measure reduced elastic modulus (Er) and hardness (H) of the coated and uncoated glass slides. Five measurements were performed in the peripheral and central parts of the slides. The AFM (Atomic Force Microscope) part and a Nanoscope E (Digital Instruments, USA) were used to obtain images of the surface. The AFM images were analyzed using Nanoscope® software version III 5.12r2. Mean roughness (Ra), root-mean square roughness (Rms), maximum height (Rmax) and 3D surface area (Rarea) were calculated using this software.
Target preparation and labeling: Genomic DNA was extracted from peripheral blood using TadbirFan Azma DNA extraction kit (Iran). For labeling genomic DNA, 2 µg of DNA was added to dH2O to bring total volume to 21 µl. Then, 20 µl of 2.5X random primer/reaction buffer was added. The mixture was boiled for 5 min, and then placed on ice. 5 µl of 10X dNTP was added on ice. Then, 3 µl of cy5-dCTP dye and 1 µl of Klenow fragment were added. The mixture was incubated at 37 ºC for 2 hr. The reaction was stopped by adding 5 µl of 0.5 M EDTA at Ph=8.0.
Hybridization: Hybridization solution was prepared according to table 2. Pre-hybridization of the slides was done to eliminate non-specific binding of target to the slides. The slides were immersed in hybridization solution and incubated at 42ºC for 1 hr in a water bath. Then, the pre-hybridized slides were washed in dH2O and dried using compressed nitrogen gas.
Labeled DNA target was diluted in hybridization solution to 10 nM final concentration. The microarrays were hybridized with appropriate amount of target under cover slips in incubator at 42ºC 5 hr until spots saturated. Then, the slides were immersed in the solution 2X SSC and 0.1% SDS and washed in 1X SSC for 1 min, in 0.2X SSC for 1 min and 0.05X SSC for 1-2 s and finally rinsed in dH2O and dried using compressed nitrogen gas.
Microarray quantification: The hybridized microarrays were scanned using Scan Array GX Microarray Scanner (Perkin Elmer). Spots were quantified from the images generated from Scan Array GX Microarray software (Perkin Elmer) using Scan Analyze software version 2.5. For calculating signal-to-noise ratio (SNR) of spots, the following formula was employed:
Where SM was mean signal of four spots which replicated each probe when hybridized to cy5 labeled target and BM was mean background signal.
Results :
The principle for linking of different coatings with biomolecules: PLL used to coat glass slides for preparing microarrays can attach to glass slides more strongly than agarose layer. But PLL-coated surfaces have low binding capacity for biomolecules. Activated agarose provided a thin layer on the glass with active aldehyde groups which can bind to amino groups on various biomolecules, such as DNA, proteins, peptides and phospholipids. In addition to providing active groups on the surface, agarose could perform a physical function; it provided a uniform three-dimensional solid support for dynamic attaching of biomolecules. Activated agarose thin film on the PLL-coated glass slides resulted in chemical binding between two polymer layers; therefore, agarose could attach to PLL-coated slides more strongly than unmodified slides. Therefore, higher stability in hybridization reactions and higher signal-to-noise ratio were provided.
Surface analysis of polymer film-coated slides: The surface profiles of the coated and uncoated glasses characterized by AFM showed that agarose provided three-dimensional thin film on the unmodified and PLL-coated glass slides (Figures 1A and 2A). The mean roughness (Ra), root-mean square roughness (Rms), maximum height (Rmax) and 3D surface area (Rarea) for agarose-PLL coated, agarose coated and PLL- coated glass were reported in table 3. The section analysis of agarose-PLL coated, agarose coated and PLL-coated slides revealed that their vertical distance were 35.520 and 33.504 nm, respectively (Figures 1C, 2C and 3C), while vertical distance of PLL-coated surface was 0.817 nm. The distance demonstrated the thickness of thin polymeric films on glass surface.
The mean reduced elastic modulus (Er) and mean hardness (H) were calculated from five indentations performed in peripheral and central parts of surface and were reported in table 4 for agarose on PLL-coated slides, agarose on unmodified glass and unmodified glass. The nano-indentation measurements on the surfaces ascertained the impressibility and softness of prepared substrate surface which was essential for spotter pins’ safety. Although, agarose coatings were applied manually to the surface, five different measurements showed that agarose layers were relatively uniform. The difference between mechanical properties of agarose-PLL in comparison with PLL two-dimensional layers was related to three-dimensional agarose structure on the surface. PLL is a polymer which can reduce hardness of the coated surface; therefore, the difference between the mechanical properties of agarose-PLL and agarose coatings and also the difference between PLL-coated and unmodified glass surfaces were related to coating by PLL polymeric layer.
Spot uniformity and morphological analysis of prepared microarrays: SEM analysis of the agarose on the glass slides showed a porous film (Figure 4). In addition, this micrograph demonstrated that the orientation of agarose film was parallel to glass surface.
The SEM micrograph of the prepared microarrays on three different coated substrates showed spots written on the agarose layers were more regular than spots on the PLL-coated slides in shape and size (Figures 5A, B and C). Some spots on the PLL-coated substrate had an inappropriate quality and were ununiformed in diameter. It is worth mentioning that spots on the agarose had better quality (Figures 6A, B and C). The spot diameter was between 200-250 µm.
Stability of polymer films in hybridization process: The activated agarose layer grafted to PLL- coated on the glass surface could sustain incubation at 37ºC for 6.5 hr and at 50ºC for 5 hr. Also, we examined the stability of agarose layer on the unmodified glass slides; it could sustain incubation at 37ºC for 3 hr and at 50ºC for 1 hr without rehydration and detaching from the slides. These slides can be stable in hybridization of biomolecules, such as proteins, peptides and oligonucleotides to microarrays.
Comparison of different microarray substrates in hybridization reactions: The slides coated using activated agarose and agarose-PLL film showed significant increase in signal and signal-to-noise ratio in comparison with slides coated using PLL alone (Figures 7A and C). Also, background signal from substrate was higher in PLL-coated slides than two slides coated using agarose. The agarose layer on PLL-coated substrate showed the best hybridization signal (Figure 7B). The agarose-PLL coated substrates had a 4-fold and 2-fold increase in SNR compared with PLL-coated slides and agarose-coated slides, respectively. These results demonstrated that agarose-PLL coated slides are suitable for analyzing wide concentration range analytes.
Discussion :
The physical methods for modification of glass substrates to immobilize biomolecules provide surfaces with low binding capacity which result in non-uniform absorption. Therefore, the probe immobilization isn’t efficient to hybridize to target molecules and insufficient signal-to-noise ratio may be obtained. Using chemical immobilization, based on covalent crosslinking of surface active groups and chemical active groups on probe structure, uniform and efficient immobilization of biomolecules on the solid substrate can be provided. Agagrose gel is considered as a non-toxic and inexpensive polysaccharide which has matrix structure and specific physical and chemical properties, such as high gel strength at low concentrations, broad range of physical, chemical and thermal stability and low degree of chemical complexity. Therefore, it has been widely used in biotechnology and life sciences, such as immobilization substrate in electrophoresis 21.
Previous studies ascertained active aldehyde groups in agarose could be linked to amino groups in biomolecules and after UV crosslinking, the covalent binds could be formed. On the other hand, agarose could attach to surface of the glass more strongly if amine groups were provided on the surface. In this research, we used a PLL coating on the glass surface which could be linked to some of the aldehyde groups in oxidized agarose. PLL is a material utilized for glass modification to prepare microarrays. This polymer can attach to glass surface strongly. So, it has low interference in background signal; also it is a polymer and can provide high density of amine groups for binding more uniformly to agarose. Because of the gel structure of agarose, thin film biomolecules could infiltrate into its microporous structure so binding capacity of surface would be increased. The surface analysis of the prepared slides demonstrated that agarose gel was a three-dimensional matrix which resulted in an increase in surface roughness and a decrease in hardness and elastic modulus compared with PLL coated slides and unmodified glass. The hardness and modulus of agarose-PLL layer were lower than agarose coating on the glass and the coating of PLL polymer on the glass substrate under the agarose thin film was shown.
In addition, these results ascertained impressibility and softness of prepared substrates that were essential for the spotter pins’ safety during contact with the surface in mechanical spotting. The agarose-PLL layer, which had the lowest hardness, had low risk of pin breakage during contact with pins’ head. The vertical distances for agarose-PLL and agarose surface were 35.520 and 33.540 nm, respectively. This parameter could estimate the thin film layer on the surface. The SEM images of the spots on three different substrates showed that spots on two types of agarose layers (agarose on PLL and agarose) were more regular in shape and size in comparison with the spots on PLL coated surface. This observation could be related to desirable hydrophilicity/hydrophobicity of agarose surface compared with PLL 21.
Long-term stability of agarose layer grafted to PLL was significantly increased compared with agarose on the surface of the other amino-modified glass 19 and agarose on the ordinary glass slides. The agarose on PLL-coated glass sustained itself for 6.5 hr at 37ºC and for 5 hr at 50ºC, whereas agarose film on unmodified glass and agarose layer on the other modified glass were stable for 1 and 4 hr 19 at 50ºC, respectively. This was the result of binding of some of the active aldehyde groups of agarose to amine groups of PLL on the surface and stronger attaching of agarose to surface of the glass substrate.
Effective binding of probes to the surface of agarose-PLL coated and agarose coated substrates reduced noise in hybridization signal compared with PLL coated substrates. Also, the stronger attachment of agarose to PLL coated glass than the unmodified glass increased signal-to-noise ratio (56.435 compared with 21.235, respectively) and decreased background signal (205 compared with 381, respectively) which resulted in sufficient hybridization analysis on the agarose-PLL coated glass substrates.
In previous related studies, researchers immobilized amino-modified probes on to agarose-coated substrates 19,22,23. Dufva et al immobilized 20-25 bp long modified probes by UV irradiation which resulted in significant increase in hybridization signal 19. In another study, this group immobilized poly(C) 10- poly (T) 10 tagged 17-base oligonucleotide, but unmodified probes, using UV light on to agarose surface and they obtained sufficient discrimination signals 20. In our research, the unmodified longer probes (35-40 bp) were immobilized on agarose surface using UV irradiation, therefore, despite the connection between few bases in probe end and the gel surface, the probes had enough number of nucleotide to retain their specificity for hybridization to targets as ascertained by sufficient signal intensity.
In addition, utilization of PLL coating under agarose layer on glass surface resulted in higher stability and lower background signal in hybridization compared with silane coating under agarose layer 19,23. This was the reason for low interference of PLL in fluorescence detection.
Conclusion :
In conclusion, we can say that substrates coated with polymeric thin films provide sufficient mechanical properties for robotic spotting, three-dimensional structure for binding unmodified oligonucleotide, low background noise, and high signal intensity in hybridization reactions.
Therefore, we fabricated inexpensive and simple to use microarray substrates, capable of biomolecule detection in high throughput and simultaneous assays.
Acknowledgement :
The authors would like to acknowledge that the study is supported by grant No. 88001818 from the Iran National Science Foundation (INSF).
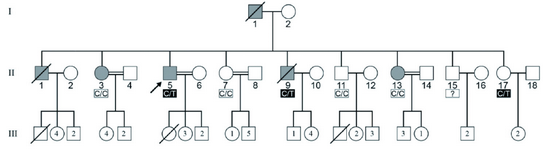
Figure 1. The pedigree of an Iranian IBGC-affected family showing transmission of autosomal dominant trait. Filled symbols represent individuals clinically and radiologically affected. The heterozygous C/T mutation was found in both the affected and unaffected members
|
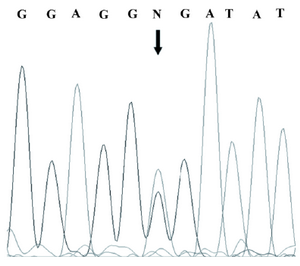
Figure 2. Sequencing result of the promoter region of the SPP2 gene in the index subject of the pedigree which showed a heterozygous C/T mutation
|
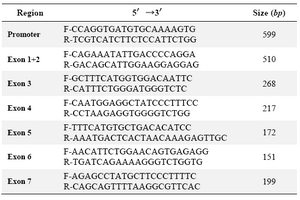
Table 1. Primer sequences for amplification of the promoter and coding region of the SPP2 gene
|
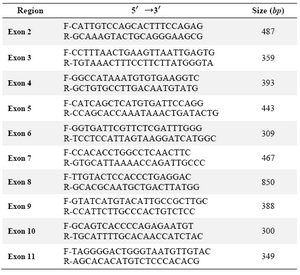
Table 2. Primer sequences for amplification of the coding region of the SLC20A2 gene
|
|