Development of One-Step Tetra-primer ARMS-PCR for Simultaneous Detection of the Angiotensin Converting Enzyme (ACE) I/D and rs4343 Gene Polymorphisms and the Correlation with CAD Patients
-
 Heidari, Mohammad Mehdi
Department of Biology, Faculty of Science, Yazd University, Yazd, Iran, Tel: +98 035 31233381, Fax: +98 035 38210644, E-mail: Heidarimm@yazd.ac.ir
Heidari, Mohammad Mehdi
Department of Biology, Faculty of Science, Yazd University, Yazd, Iran, Tel: +98 035 31233381, Fax: +98 035 38210644, E-mail: Heidarimm@yazd.ac.ir
-
Department of Biology, Faculty of Science, Yazd University, Yazd, Iran
-
Hadadzadeh, Mehdi
-
Department of Cardiac Surgery, Afshar Hospital, Shahid Sadoughi University of Medical Sciences, Yazd, Iran
-
Fallahzadeh, Hossein
-
Research Center of Prevention and Epidemiology of Non-Communicable Disease, Shahid Sadoughi University of Medical Sciences, Yazd, Iran
Abstract: Background: The Angiotensin Converting Enzyme (ACE) Insertion/Deletion and rs4343 gene polymorphisms could be associated with pathogenesis of essential hypertension and cardiovascular disorders and Coronary Artery Disease (CAD). In the present study, a fast and novel approach of multiplex Tetra-Primer Amplification Refractory Mutation System-PCR (T-ARMS-PCR) was developed for simultaneous detection of two SNPs including ACE I/D (rs4340) and 2350A>G (rs4343) of Angiotensin Converting Enzyme (ACE) gene.
Methods: The present research was performed using 148 blood samples taken from patients with CAD and 135 healthy individuals. One set of inner primers (for rs4343) and one set of outer primer pairs were designed for genotyping of Insertion/Deletion and rs4343 polymorphisms in single tube T-ARMS-PCR.
Results: Our results manifested that genotypes and alleles frequency of the ACE polymorphisms showed no statistically significant association between CAD patients and the control group. In addition, complete concordance was seen between sensitive Tetra-ARMS-PCR and sequencing method.
Conclusion: The technique is the first work for simultaneous detection of Insertion/Deletion polymorphism and rs4343 SNPs in ACE gene and the results were entirely according to those from an independent procedure.
Introduction :
Coronary Artery Disease (CAD) is the result of atherosclerotic occlusion of the coronary arteries. Most of all deaths in the developing world are due to Cardiovascular Disease such as hypertension and the diseases caused by atherosclerosis 1. Various genetic variations are known to affect coronary atherosclerotic plaques 2-4.
A key physiological regulator of blood pressure is the Renin-Angiotensin System (RAS). An enzyme which determined the vasoactive peptide Angiotensin-II in RAS pathway is angiotensin converting enzyme (ACE or Peptidyl-Dipeptidase A). The most common polymorphism in ACE gene is a 287-base-pair Insertion/Deletion (I/D) Alu element within the intron 16 of gene (rs4340 SNP). It was found that this polymorphism is related to the ACE activity level which is drastically reduced in homozygous deletion carriers (DD) when compared to homozygous II carries, while heterozygous ID carriers show middle activity level 5. Another single nucleotide polymorphism in ACE gene is a silent and synonymous coding polymorphism 2350A>G (rs4343) in exon 17. The association between ACE I/D (rs4340) and 2350A>G (rs4343) polymorphisms has been found in several studies including high blood pressure, systemic lupus erythematous, CAD, diabetic nephropathy, Alzheimer’s disease and renal diseases 6,7.
In this study, a case-control evaluation of ACE polymorphisms was done to examine their putative association with CAD in an Iranian population. Since the distance between the ACE I/D (rs4340) and 2350A>G (rs4343) polymorphisms is 139 bp, a new Tetra primer-amplification refractory mutation system-PCR (T- ARMS-PCR) method was developed for simultaneous detection of these SNPs in single reaction.
Materials and Methods :
Patients: The present study was a case-control association study. Peripheral blood samples were collected from 283 subjects (148 CAD patients and 135 age and sex matched healthy controls with no family history of CAD) who referred to cardiac centers in Afshar Hospital (Yazd, Iran) from 2012 to 2015. CAD patients were selected according to the coronary angiography and Electrocardiogram (ECG) criteria 8. Routine biochemical measurements were obtained from patients with CAD and healthy subjects including determination of fasting lipid levels High-Density Lipoprotein (HDL), triglycerides, and Low-Density Lipoprotein (LDL) levels (Table 1).
The control group consisted of 135 unrelated healthy age, sex and ethnicity matched subjects with normal or near- normal angiography results. Our protocol was approved by Yazd University Human Research Ethics Committee, in accordance with the revised declaration of Helsinki and all participants gave their informed consents for the molecular analysis. Genomic DNA was extracted from the whole blood by a salting out method.
Genotyping: All primers used in this research were designed by a web primer design program, Primer1, accessible at http://primer1.soton.ac.uk/primer1.html (Table 2). The PCR reaction was carried out in a total volume of 25 µl, containing 50 ng of genomic DNA as template, 5 pmol of each outer primer, 10 pmol of each inner primer, and 1X multiplex PCR master mix (Yekta Tajhiz Azma Co., Tehran, Iran). PCR amplification (Touchdown) was performed with an initial denaturation at 95ºC for 2 min, followed by denaturation at 95oC for 20 s, first annealing at 69oC to 60ºC (10 cycles) and the remaining cycles (25 cycles) were carried out with annealing at 60oC for 1 min and extension at 72ºC for 1 min, followed by a final extension for 5 min. The amplified products were run on non-denaturing polyacrylamide gel electrophoresis (6%) and were stained with silver staining method.
The ACE I/D (rs4340) and 2350A>G (rs4343) polymorphisms are located in intron 16 and exon 17, respectively, with 139 bp distance. Four primers were used in the same PCR. Two primers, Fo and Ro were designed to amplify a 712 bp band (I allele) and 424 bp band (D allele) which served as control bands for the success of the amplification (Figure 1). Two specific primers, Fi and Ri for rs4343 with complementary 3'-end nucleotide to corresponding polymorphisms, were introduced (Table 2).
Statistical analysis: The Chi-square goodness-of-fit test was used to investigate the association between the two groups. Values of p<0.05 were considered statistically significant. Statistical package for the social sciences (SPSS) software (IBM SPSS 22, SPSS Inc., Chicago, IL., USA) was used to perform the statistical analysis.
Results :
The average serum levels of cholesterol and triglycerides were comparable between cases and controls, whereas their levels were higher in the CAD patients (p<0.05). However, no significance differences were observed in the LDL and HDL levels between both groups (Table 1). Both of means in systolic and diastolic blood pressure showed important statistical differences in patients and controls (p<0.05).
The simple Tetra-ARMS-PCR method can determine the arrangement of alleles on chromosome. This arrangement means that insertion allele (ACE I allele) and A allele (in rs4343) are found on one chromosome (IA), which is referred to as the coupling, or Cis configuration. Alternatively, one chromosome might bear the deletion allele (ACE D allele) and A allele (DA) which is called the repulsion, or Trans configuration. The identification of each genotype arrangement (Cis or Trans state) was performed by comparing to the expected fragment lengths (Table 3). PCR products with these allele arrangements including IA arrangement were: 712 and 271 bp fragments, IG arrangement: 712 and 488 bp fragments, DA arrangement: 712 and 271 bp fragments and DG alleles: 424 and 200 bp fragments (Figure 2).
The allelic discrimination for all patients with CAD and normal samples was performed through T-ARMS-PCR by two outer primers in the presence of two inner specific primers for ACE polymorphisms. Furthermore, genotypes of a limited number of samples were analyzed through direct sequencing (for verification). It was found that genotyping results obtained from both methods were fully concordant. Two types of polymorphism in the ACE gene, ACE I/D (rs4340) and 2350A>G (rs4343) were analyzed and their genotype distributions and allele frequencies in CAD patients and healthy subjects are shown in table 4.
All the patients and controls were genotyped for rs4340 and rs4343 polymorphisms by logistic regression analysis according to co-dominant, dominant and recessive models (p>0.05). Our result showed that there was no significant difference in ACE polymorphisms genotype distribution and allele frequency between CAD patients and the control group. Table 5 shows the results of the haplotype analysis.
Discussion :
Numerous experimental studies have suggested that ACE is a key enzyme in the production of angiotensin II in the modulation of cardiac growth. Genotyping of ACE gene polymorphisms has provided a genetic marker for several human heart diseases such as ischemic heart disease, coronary artery stenosis, myocardial infarction and ischemic cerebrovascular disease 9,10. Association of these markers such ACE I/D (rs4340) and 2350A>G (rs4343) polymorphisms has been found previously by researchers in different populations (Table 6). Differences in various populations are due to variations in genetic and environmental background.
The standard PCR instruments are highly desirable for scientific studies of large number of patients and for diagnostic analysis, economical and fast assays. For conventional genotyping of ACE gene polymorphisms, scientists developed PCR-based methods which allow discrimination of different ACE genotypes. For diagnostic analysis in large numbers of patients, fast and acute assays that can be carried out with standard PCR equipment are highly desirable.
In this research, a rapid, reliable, sensitive and easy to use assay for detecting ACE I/D (rs4340) and 2350A>G (rs4343) polymorphisms in our population was developed and compared with reported results in different investigations. A one-step Tetra-primer ARMS- PCR assay was designed and genotype results were obtained that were fully concordant with direct sequencing of randomly selected samples (n=18). Furthermore, one-step Tetra-primer ARMS- PCR assay is easy to perform and needs only a small amount of standard PCR reagents and is applicable to identify the genetic variations without special equipment.
For the first time, Saddick et al used a novel approach of tetra primer ARMS-PCR for genotyping ACE I/D (rs4340) polymorphism in pregnant women with Mild Gestational Hyperglycemia (MGH) 11. In this manner, simultaneous distinction of homozygous and heterozygous samples for ACE I/D (rs4340) and 2350A>G (rs4343) polymorphisms was achieved by tetra primer ARMS-PCR. Our data showed that none of these polymorphisms was associated with CAD in Iranian patients. There were no statistically important associations between genotype distribution, allele and haplotype frequencies between cases and healthy controls. Our results are in agreement with some previous research such as Poorgholi et al and Ancelin et al, who did not find any association between ACE I/D (rs4340) and 2350A>G (rs4343) polymorphisms and CAD development 12,13. These results show that cardiovascular disorders may be caused not only by angiographic characteristics but also by combination of some genetic factors associated with environmental risk factors. Moreover, the disagreement between reported results could be not only related to difference in ethnicity and sample size, but also connected to differences in clinical presentation, severity of disorders, age and gene-environment interactions.
Conclusion :
In conclusion, it should be noted that a simultaneous detection method for two different variations in ACE gene was presented by one-step Tetra-primer ARMS- PCR assay. However, our study does not report a significant association of ACE I/D (rs4340) and 2350A>G (rs4343) polymorphisms in Iranian patients with coronary artery disease. Further researches are required to completely understand the role ACE variations in different populations.
Acknowledgement :
This research was funded by Yazd University. We thank all the patients for providing blood samples for the scientific research, also, the Especial Afshar Hospital (Yazd, Iran).
Conflict of Interest :
All authors declare no conflicts of interest.
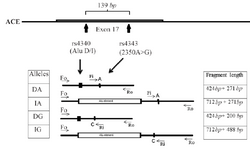
Figure 1. Schematic illustration of primer interactions for ACE Multiplex T-ARMS-PCR assay. Fo and Ro are outer primers acting as control primers, and Fi and Ri are inner primers for 2350 nucleotide substitution (rs4343).
|
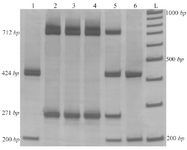
Figure 2. Results of T-ARMS-PCR of ACE polymorphisms (rs4043 and rs4343). Lanes 1 and 6: DDGG genotype, lanes 2, 3 and 4: IIAA genotype and lane 5: DIAG (IA/DG) genotype.
|
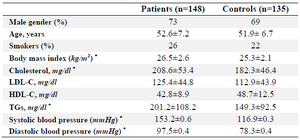
Table 1. The summary of the clinical characteristics of coronary atherosclerosis patients and controls
* p-value <0.05
|

Table 2. PCR primer sequences
|
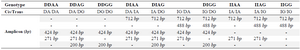
Table 3. Amplification patterns observed and those expected according to ACE polymorphisms genotypes
D and I: Deletion/ Insertion allele (rs4340 SNP); A and G: wild type and mutant allele (rs4343 SNP).
|
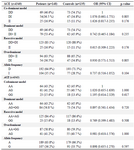
Table 4. Genotype and allele frequencies of ACE variants in patients and controls
OR, odds ratio; CI, Confidence interval.
|
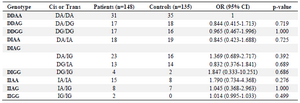
Table 5. Haplotype analysis of AGT polymorphisms in CAD patients and controls
OR, odds ratio; CI, Confidence interval.
|
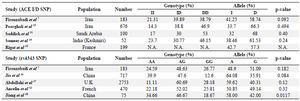
Table 6. The results of different studies on ACE gene polymorphisms
|
|