Comparison of Epothilone and Taxol Binding in Yeast Tubulin using Molecular Modeling
-
Akbari, Vajihe
-
Department of Pharmaceutical Biotechnology, Faculty of Pharmacy, Isfahan University of Medical Science, Isfahan, Iran
-
Moghim, Sharareh
-
Department of Bacteriology and Virology, Faculty of Medicine, Isfahan University of medical sciences, Isfahan, Iran
-
 Mofid, Mohammad Reza
Department of Biochemistry, School of Pharmacy and Pharmaceutical Science, and Bioinformatics Research Center, Isfahan University of Medical Sciences, Isfahan, Iran, Tel: +98 311 7922597 Fax: +98 311 6680011 E-mail: mofid@pharm.mui.ac.ir
Mofid, Mohammad Reza
Department of Biochemistry, School of Pharmacy and Pharmaceutical Science, and Bioinformatics Research Center, Isfahan University of Medical Sciences, Isfahan, Iran, Tel: +98 311 7922597 Fax: +98 311 6680011 E-mail: mofid@pharm.mui.ac.ir
-
Department of Biochemistry, School of Pharmacy and Isfahan Pharmaceutical Sciences Research Center, Isfahan University of Medical Sciences, Isfahan, Iran
Abstract: Microtubules are unique cytoskeletal structures that have structural subunits of αβ tubulin. Taxol is a typical microtubule stabilizing drug. The epothilones are other natural products with similar mechanism of action totaxol. Despite the highly conserved nature of β-tubulin, some organism like Saccharomyces cerevesia is resistance to taxol, but sensitive to epothilones. In order to find differences in sensitivity of yeast tubulin to these molecules, we investigated binding mode of the taxol and epothilone A to yeast tubulin using molecular modeling. The multiple sequence alignment of β-tubulin of different species was performed using ClustalW2. Protein structure of yeast β-tubulin was constructed with Swiss Model 8.05 by using 1TVK. Modeled tubulin was superimposed with PyMol on1JFF for comparison of three-dimensional structure of two proteins. Our results showed that one of the most interesting differences in binding mode of these molecules is residue 227. The His227 in bovine makes a hydrogen bond by means of its δ-nitrogen with epothilone A and by means of its ε-nitrogen with taxol. The Asn227 of yeast can play role of the δ-nitrogen of imidazole ring of H227, but not of ε-nitrogen of it. So yeast tubulin in contrast to taxol can interact with epothilone A. Due to conservation of essential residues for binding (T274, R282 and Q292), epothilone A in comparison with taxol can tolerate the interchange in the binding pocket (R276I). Our findings may be of a great aid in the rational design of anti-tumor agents that bind to the taxol binding region of tubulin.
Introduction :
Microtubules are unique cytoskeletal structures that have structural subunits of αβ tubulin, which are important for intracellular transport and cell division of all eukaryotes (1). An effective and validated target for tumor chemotherapeutic agents is the microtubule cytoskeleton. These anticancer drugs can be divided into two main classes: those that inhibit microtubules assembly and those that stabilize microtubules. Taxol is a typical microtubule stabilizing drug of the first group. Other natural products with the similar mechanism of action to taxol including epothilones, discodermolide, eleutherobin, sarcadictyin and laulimalides, have been isolated and are in various stages of preclinical/clinical development (2).
In experiment with bovine brain tubulin, epothilones competitively inhibit the binding of taxol to microtubules (3,4). This data suggested that there is a common binding site or overlapping site for two compounds, despite having unrelated structures. In comparison with taxol, epothilones are more effective than taxol against some multidrug resistance cell lines and are more water soluble than taxol (3).
Despite the highly conserved nature of β-tubulin across the phyla, organisms show different degrees of susceptibility and resistance to the different groups of antimicrotubule drugs. For example, many protist and fungal tubulins are only slightly affected by taxol (5). However, they are structurally normal and can even co-assemble with mammalian tubulins (6). It has been reported that sensitivity to this molecule is controlled by relatively few residues.One study (7) made a taxol-binding site in yeast β-tubulin, which is normally taxol-resistant, by replacing five residues with their mammalian counterparts (A19K, T23V, G26D, N227H, and Y270F).
According to the high degree of conservation among tubulin proteins and the relative simplicity of the yeast microtubule cytoskeleton, Saccharomyces cerevesia (S.cerevesia) is a suitable organism for studying the microtubule cytoskeleton. Tubulin of the budding yeast S.cerevesia shares 76% sequence identity with bovine tubulin, but yeast tubulin isn't sensitive to taxol. In contrast, epothilones can promote the assembly and stabilization of yeast microtubules. So the sequence and structure differ in yeast tubulin and bovine brain tubulin that allow strong binding of epothilones, but not of taxol (8-11).
In view of the fact that S.cerevesia has only one gene (TUB2) encoding β-tubulin, this organism was chosen for our study. In our studies, by using molecular modeling we determined the differences in binding interactions with yeast β-tubulin among the microtubule-stabilizing agents, taxol and epothilones. Understanding of the binding mode shared by taxol and other agents with similar taxol-like activity (such as epothilones) will be a great aid in the rational design of anti-tumor agents that bind to the taxol binding region of tubulin.
Materials & Methods :
Multiple sequence alignment
Protein sequence alignments and comparisons were performed using a Basic Local Alignment Search Tool (BLAST) program, blastp, against database specification of non-redundant proteins which were available at the National Center for Biotechnology Information (NCBI) Web server, [http://www.ncbi.nih.gov/blast/] (12). Amino acid sequences were obtained from NCBI Web server, [http://www.ncbi. nlm.nih.gov/]. Multiple sequence alignments were performed using ClustalW2 [http://www.ebi.ac.uk/clustalw/], available at the European Bioinformatics Institute (EBI) Web server (13). Consensus of amino acid sequence was obtained from Boxshade available at the European Molecular Biology Network (EMBnet) Web server[http: //www.ch.embnet.org/].
Known 3D structure of proteins
Three-dimensional protein structures of bovine brain tubulin in complex with taxol (1JFF) and epothiloneA (1TVK) were obtained from the Protein Data Bank (PDB) [http//www.rcsb.org/pdb/] (14,15).
Molecular modeling
Protein structure of yeast β tubulin was constructed with Swiss Model 8.05[http:// www.expasy.org/swissmodel/] (16-19) by using a bovine brain tubulin in complex with epothiloneA (1TVK). The sequence identity between yeast β tubulin and bovine brain tubulin are 76%. The yeast tubulin structure model was quite similar to the bovine brain tubulin crystal structure. The Root Mean Square Deviation (RMSD) for 1704 polypeptide backbone atoms was 0.11 Å as calculated with Swiss-Pdb Viewer version 4.0.1[http://www.expasy.org/spdbv/] (20). We used QMEAN Z-score as an estimate of the model quality. The reliability of the modeled structure was checked by PROCHECK software online.
Molecular modeling visualization and analysis
Modeled yeast tubulin was superimposed with PyMol on the bovine β-tubulin in complex with taxol (1JFF) for comparison of three-dimensional structure of two proteins. Structural presentation of 1JFF and 1TVK and modeled yeast tubulin was made by using Swiss-Pdb Viewer and PyMOL (Version 0.99) [http://pymol.sourceforge.net] programs (21). Cartoon drawings of the structures were obtained using PyMOL (21). Studies and comparisons of taxol and epothiloneA binding interaction with yeast tubulin were performed by using programs, Swiss-Pdb Viewer and PyMOL.
Results :
The multiple sequence alignment of β-tubulin of different species is shown in figure 1. Consensus sequences obtained from the Boxshade program is shown. According to this alignment β-tubulin is highly conserved in different eukaryote.
Tubulin has binding site for GTP that highly conserved among the species. Tubulin like other nucleotide-binding proteins belongs to a class of regulatory proteins called G proteins. Consensus sequence GQC, KGHYTEG, DNEAL and glycine rich sequences SLGGGTGSGMG are binding site for phosphoryl group of nucleotide and are observed in other nucleotide binding proteins (Figure 1) (22). Residues in the 15-27, 212-231, 270-285 and 358-372 region of β-tubulin form hydrophobic sites for taxol (23) and residues in the 212-231 and 270-282 regions make contact with epothilone (24). In these regions, we observed some differences between yeast β-tubulin and β-tubulin of other species. For example, the yeast tubulin instead of K19, V23, D26, H227, A231, F270, R276 and R359 has A, T, G, N, S, Y, I and Q respectively (Figure 1).
The modeled yeast β-tubulin superimposing on the bovine β-tubulin with a RMSD of 0.11 Å and E-value of 4.2e-45 showed that the overall structure is similar (Figure 2). The QMEAN Z-score directly shows how many standard deviations the model's QMEAN score differs from expected values for native or experimental structures (e.g. unexpected solvent accessibility, back-bone geometry, inter-atomic packing, etc.) (25).Good models reach QMEAN Z-scores comparable to experimental structures (0Analysis of binding interaction of bovine β-tubulin and taxol reveals that 17 residues from the hydrophobic pocket of taxol binding site made direct contact with toxol molecule including V23, D26, L215, L217, H227, L228, A231, S234, F270, P272, L273, T274, S275, R276, P358, R359, G360 (Figure 4A).
Seven of these residues are different in yeast β-tubulin (Val23Thr, Asp26Gly, His227Asn, Ala231Ser, Phe270Tyr, Arg276Ile, Arg359Gln replacement) (Figure 4B).
Binding mode analysis of bovine β-tubulin and epothilone A showed that 6 residue from the epothilone A binding site made direct contact with epothilon A molecule including H227, A231, T274, R276, R282,Q292 (Figure 4C). Yeast β-tubulin instead of H227, A231 and R276 has N, S and I respectively (Figure 4D).
Molecular modeling shows that another important residue for binding of taxol and epothilone is position 227. Mode of interaction of taxol and epothilone with this residue is compared in figure 5. This comparison shows that these molecules have different mode of interaction with this residue.
Discussion :
Our alignment showed the high identity and homology in sequence of β-tubulin of different species. We didn't see any gap except in C terminal. At C-terminal of β-tubulin, we observed extensive molecular heterogeneity. These C terminal regions are highly variable among the isotype within a species and among species. This region is important for anchoring Microtubule Associated Protein (MAP) in place (27). The diversity of C- terminal may propose the mechanism for isotype-specific MAP binding.
Molecular modeling shows that M loop of yeast β-tubulin in comparison with bovine β-tubulin locates far away taxol molecule. M loop is the main component in contacts between protofilaments, interacting with structures in the H1-S2 and H2-S3 loops of the adjacent monomer (28). S.cerevesia exhibit some nonconservative substitutions in the M-loop compared to other species (Figure 1). The interesting substitution is at position 276. The residue R276 in the M loop of bovine β-tubulin, which is a positively charged residue, is replaced by an isoleusine in yeast. This position exists near a predicted bend in the M-loop, and an isoleusine residue here may change the shape of M-loop and its interaction with taxol. Because of the flexible long side chain and its surface location, arginine can interact with taxol.
There are many oxygen groups in taxol molecule that can interact and make hydrogen bond with positively charged goanidino group of arginine (Figure 3a). This interaction can stabilize and keep the molecule of taxol in its hydrophobic pocket. In contrast, neutral isoleusine cannot make such hydrogen bond and thus cannot stabilize taxol in its pocket binding (Figure 3b). The binding of epothilone A do not affect by this change so the yeast β-tubulin is yet sensitive to this drug. The epothiloneA molecule can interact with yeast tubulin because of existence of important hydrogen bonds between epothilone A and yeast tubulin (Figures 3C and 3D).
The isopropyl group of V23 is near the C3´-benzamidophenyl ring in the bovine β-tubulin (Figure 4A) (29). Yeast β-tubulin has a theronine residue instead of valine. Substitution of non-polar valin by polar threonine leads to distort the positioning of the hydrophobic phenyl ring (Figure 4B). In the bovine β-tubulin, the methylene group of K19, E22, D26 and E27 interact with the C3´-benzamidophenyl ring and stabilize it (28). In yeast β-tubulin, replacement of K19 and D26 by alanin and glycin respectively reduce the methylene groups that interact with the C3´-benzamidophenyl ring. The size of epothilone A molecule is smaller than taxol molecule so epothilone A only fill half of taxol binding pocket in β-tubulin. Thus, residues in 15-27 region of β-tubulin including K19, V23 and D26 which interact with taxol, do not play an important role in epothilone binding (Figure 4C) (29). We measured distance between hydroxyl group of T23 of yeast β tubulin predicted model and the C3´-benzamidophenyl ring of taxol molecule, and distance between OH group of T23 of yeast modeled β tubulin and thiazol ring of epothilone. These distances were 5.7 Å for taxol and 9.88 Å for epotilone A. So the polarity of hydroxyl group of threonine can affect the binding of taxol, but not of epothilone A.
In a mutagenesis study, Himes et al, reported that epothilones despite taxol can tolerate the change made at position 227 and bind equally well to wide-type (N227) and the mutated (H227) yeast tubulin (7).
According to the model, the imidazol ring of H227 of bovine β-tubulin is situated between C2-benzoyl phenyl and C3´-benzamidophenyl ring of taxol molecule (Figure 4A). The ε-nitrogen of imidzol ring of histidine 227 can make hydrogen bond with oxygen atom of C3´ benzamidophenyl ring of taxol, and thus stabilizes this drug in its hydrophobic pocket (Figure 5A). The thiazol ring of epothilone A and the imidazol ring of H227 of bovine β-tubulin are located in a face-to-face orientation (Figure 5B) (30). The δ-nitrogen of imidazole ring of H227 can make hydrogen bond with nitrogen atom of thiazol ring of epothilone A (Figure 5B).
In our yeast β-tubulin model, asparagine was situated position 227 (Figure 5A) and because of its distance, δ-nitrogen atom of N227 cannot play the role of ε-nitrogen of imidzol ring of H227, thus eliminates the hydrogen bonds (Figure 5A). In contrast, the δ-nitrogen atom of N227 in yeast tubulin can mimic the role of δ-nitrogen atom of histidine and makes hydrogen bond with thiazol ring of epothilone A (Figure 5B).
According to this model, the residue F270 in bovine β-tubulin was located in a hydrophobic basin (Figure 4A) (28). This residue can make van der Waals contact with C3´-phenyl ring and C4- acetyl group of taxol molecule. The epoxide ring at position C12, C13 of epothilone A accommodates the C3´-phenyl ring and C4- acetyl group of taxol (31), and is located at the entrance of the hydrophobic basin, but in contrast totaxol, it doesn’t enter this pocket. The measured distance between the C3´-phenyl ring of taxol and F270 was 3.8 Å. This distance for epoxide ring of epothilone A and same residue was 8.96 Å. In our yeast β-tubulin model, tyrosine and serine were located at positions 231 and 270. These hydrophilic side chains distort the van der Waals contacts of hydrophobic basin with C3´-phenyl ring and C4- acetyl group of taxol molecule. The residue F270 is a key residue for taxol binding, but not to the epothilone A. So the change of this residue in contrast totaxol does not affect the binding of epothilone to yeast β-tubulin. The cytotoxicity data show that the Phe270Val mutation has only a 3-fold effect on the sensitivity of the epothilone A compared with a 27-fold change for the taxol (32), confirm this finding.
The position 359 in bovine β-tubulin is occupied by arginine that makes hydrogen bond with taxol molecule (Figure 4A). In yeast β-tubulin glutamine locates at position 359 and distorts this hydrogen bonding. In agreement with our finding, Henriquezet al reported that sequence replacement of Arg359Ala in Acanthamoeba tubulin lead to resistance to taxol (33). The distances of this residue with taxol and epothilone A was measured 3.2 and 8.4 Å, respectively. According to this finding, epothilone is so far to make direct hydrogen bound with this residue so that its binding is not affected by changing this residue.
According to the model, the 7-OH group of epothilone and oxetane ring of taxol made hydrogen bonding with T274. The residue T274 is important for binding of both of taxol and epothilone A and it is confirmed by mutagenesis study which reported that a Thr274Ile replacement had a determined effect on activity (34). This effect is more emphasized on epothilone than on taxol activity. This residue is conserved in yeast so the epothilone A can made one of the most important interaction with its β-tubulin.
The model also showed that residue arginine at position 282 is essential for binding of epothilone (Figure 4B), but not of taxol. Again, this residue is conserved in yeast thereby epothilone A can bind to its β-tubulin. This positively charged residue made electrostatic contacts with the 7-OH group of epothilone A. On the other hand, R282 can form hydrogen binding with T274 and thereby stabilizing conformation of M loop for interaction with epothilone A. In contrast, taxol doesn’t make such network of electrostatic interactions and this residue isn’t a key residue for its binding and again mutagenesis study agrees with this fact (24).
The other residue that is important for binding of epothilone, but not to the taxol is Q292 and again this residue is conserved in yeast, so this organism is sensitive to epothilone A (Figure 4B). This residue locates near the M loop from and forms hydrogen binding with Leu273 adjacent to T274 (15). Mutagenesis studies showed that Gln292Glu replacement distorts this hydrogen bonds network of M loop so inhibits epothilone binding (34).
Conclusion :
Analysis of S.cerevesia β-tubulin has shown that failure in interaction with the C3´-benzamidophenyl and phenyl rings and stabilizing them, because of Val23Thr, Asp26Gly, His227Asn, Ala231Ser, Phe270Tyr replacements, is the main reason for resistance to taxol. Study of modeled yeast tubulin revealed that ability of epothilone to stabilize yeast tubulin is related to conservation of key residues for epothilone binding (including T274, R282 and Q292) in this organism.On the other hand, epothilone A can tolerate some interchange in taxol binding pocket residues due to flexibility of epothilone A in comparison with taxol.
Our direct comparison of epothilone binding and taxol binding in yeast tubulin can support development of novel natural and synthetic microtubule stabilizing agents with higher affinity for tubulin and lower sensitivity toward development of resistance of cancerous cells.
This would lead to make anticancer drugs with an improved efficiency and fewer side effects.
Acknowledgement :
Special thanks are extended to Dr. Ahmad Reza Shahverdi and Dr. Mojtaba Panjehpour for their helpful suggestions. This research was supported by the Bioinformatics Research Center of Isfahan University of Medical Sciences.
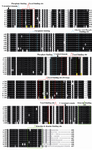
Figure 1. Multiple alignment of β-tubulin of different spe-cies: a) Oryza sativa, b) Plasmodium falciparum, c) Salmo-salar, d) Bostaurus, e) Drosophila melanogaster, f) Homo sapiens, g) Cyathostomumcatinatum, h) Gallus gallus and i) Saccharomyces cerevesia. Key residues for taxol binding (red), epothilone A binding (orange) and common residues for binding of taxol and epothilone A (green) are highlighted
|
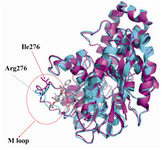
Figure 2. Superimposition of backbone atoms of modeled yeast β-tubulin (light magenta) on bovine β-tubulin (cyan). Arrows indicates the position of the M loop and residue 276 in the β-tubulin
|
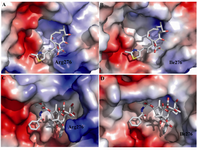
Figure 3. A) Residue 276 of bovine tubulin and its interaction with taxol, B) Residue 276 of yeast tubulin and its interaction with taxol, C) Residue 276 of yeast tubulin and its interaction with epothilone A and D) Residue 276 of bovine tubulin and its interaction with epothilone A
|
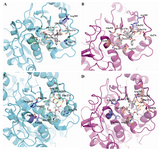
Figure 4.Comparison of the binding mode of paclitaxel and epothilones in yeast and bovine tubulin. Ligands (gray), binding-site key residues (greencyan) and non-key residues (slate) for bovine tubuline, and), binding-site key residues (salmon) and non-key residues (slate) for yeast tubuline that e are shown in stick representation. Hydrogen bonds are presented as yellow long dashes and hydrophobic interactions are shown as red square dots. The remaining part of the β-tubulin secondary structure is rendered with cartoon presentation using PyMOL. A) Taxol as found in the electron crystallography model (PDB: 1JFF). B) Taxol that interacts with modeled yeast tubulin. C) Epothilone A as found in the electron crystallography model (1TVK). D) Epothilone A that interacts with modeled yeast tubulin
|
![Figure 5. Possibility of hydrogen bonding between residue 227 of tubulin [Histidine in bovine (cyan) and Asparagine in yeast (magenta)] and A) oxygen atom of 3´-benzamidophenyl group of taxol, B) nitrogen atom of thiazol ring of epothilone A](Images/Articles/73/f5_small.png)
Figure 5. Possibility of hydrogen bonding between residue 227 of tubulin [Histidine in bovine (cyan) and Asparagine in yeast (magenta)] and A) oxygen atom of 3´-benzamidophenyl group of taxol, B) nitrogen atom of thiazol ring of epothilone A
|
|