Recombinant Production of TP4-LYC1, A New Chimeric Peptide with Targeted Cytotoxicity to HeLa Cells
-
Mohammad Pour, Hanieh
-
Department of Pharmaceutical Biotechnology, School of Pharmacy and Pharmaceutical Sciences, Isfahan University of Medical Sciences, Isfahan, Iran
-
Jahanian-Najafabadi, Ali
-
Department of Pharmaceutical Biotechnology, School of Pharmacy and Pharmaceutical Sciences, Isfahan University of Medical Sciences, Isfahan, Iran
-
 Shafiee, Fatemeh
Department of Pharmaceutical Biotechnology, School of Pharmacy and Pharmaceutical Sciences, Isfahan University of Medical Sciences, Isfahan, Iran, Tel: +98 313 7927058; Fax: +98 313 6680011; E-mail: f_shafiee@pharm.mui.ac.ir
Shafiee, Fatemeh
Department of Pharmaceutical Biotechnology, School of Pharmacy and Pharmaceutical Sciences, Isfahan University of Medical Sciences, Isfahan, Iran, Tel: +98 313 7927058; Fax: +98 313 6680011; E-mail: f_shafiee@pharm.mui.ac.ir
Abstract: Background: Tilapia Piscidin 4 (TP4) showed potential anti-tumor effects against various cancer cells. Lycosine-1 (LYC1), is another Antimicrobial Peptides (AMP) from spider venom with targeted penetration to cancer cells without any adverse effects on normal cells. The aim of this study was to produce a soluble recombinant fusion peptide in order to diminish the cytotoxicity of TP4 against normal cells.
Methods: In order to express of TP4-LYC-1, TP4, and LYC1 in fusion to the inteins1/2 of pTWIN-1 vector, induction condition was optimized to earn soluble peptides. Auto-cleavage induction of inteins1/2 was performed based on IMPACT® manual and their effect on cell viability of HeLa and HUVEC cells was surveyed by MTT assay.
Results: The best condition for accessing the most soluble peptide in fusion to the inteins was approximately similar for all three peptides (0.1 mM of IPTG, at 22°C). After the induction of self-cleavage of inteins, a band in 3, 3, and 6 kDa was observed on tricine-SDS-PAGE. The IC50 values of TP4-LYC1 and TP4 against HeLa cells were calculated as 0.83, and 2.75 µM, respectively.
Conclusion: In the present study, a novel chimeric peptide, TP4-LYC1, was successfully produced. This fusion protein can act as a safe bio-molecule with potent cytotoxic effects against cancer cells, but the penetration ability and determination of cell death mechanism must be performed in order to have more precise view on the apoptosis induction of this recombinant peptide.
Introduction :
Cationic Antimicrobial Peptides (AMPs) are natural biological agents with numerous medicinal potentials. These peptides have been identified and isolated from various sources such as insects, fishes, amphibians, and animals with an important role in innate immunity against microbial pathogens 1,2. Some specific types of these peptides, such as secropin, have also showed good anti-cancer effects on human cancers 3.
In comparison to the traditional chemotherapeutic agents, these peptides have shown many advantages; most of these peptides do not have significant cytotoxic effects on normal and non-proliferating cells at therapeutic concentrations 4. Therefore, in vivo use of these agents is expected to be associated with minimal non-specific toxicity 5. Another important advantage of these peptides is the lack of drug resistance compared to chemotherapy drugs 6. Most AMPs also have the ability to inhibit angiogenesis 7 with a high potential to act as anticancer agents. Finally, their regulatory effects on the immune system are the other suggested mechanism to act as anti-tumor agents 8. So, using these agents creates a new and progressive opportunity for the cancer treatment.
One AMP with anticancer effects on various cancer cells is TP4 which is extracted from the Nile tilapia (Oreochromis niloticus) with 25 amino acids residues and an alpha helix structure 9. The cytotoxic and apoptotic effects of this peptide have been evaluated in several studies. For example, a study has shown that this peptide penetrates into the cells, targets the microtubule network, prevents microtubule polymerization and consequently, kills cancer cells. On the other hand, after the treatment of A549 cells with this peptide, the microtubule network was disrupted and cell death was induced 10.
Another study by Ting et al showed that this peptide exerts cytotoxic effects on lung cancer cells including A549, NCI-H661, NCI-H1975, and HCC827 through the induction of necrosis in these cells 11. The effects of this anticancer peptide on breast cancer cell lines have also been studied; it was found that TP4 induces necrosis in MDA-MB231, MDA-MB453, and MCF7 cells 12.
Finally, in the Su et al’s study, the necrotic effects of this peptide on glioblastoma cells have been proven. In the mentioned study, it was shown that TP4 induces cell death through necrosis in the U87MG and U251 cell lines and releases cyclophilin A as a marker of necrosis, as well as leading to mitochondrial dysfunction and increased levels of oxygen free radicals in cells 13.
However, another recent study has shown that this peptide induces apoptosis in MG-63 osteosarcoma cells in approximately 10 times lower concentrations than the concentrations used in the previous study, which led to apoptosis in cancer cells 14. Therefore, the use of this peptide can be a good treatment for different types of cancer.
On the other hand, the use of vehicles that can transfer the cytotoxic agents only to cancer cells in order to minimize their entry and toxicity in normal cells, helps to target cytotoxic agents and reduce the final therapeutic dose. In this study, LYC1 (Lycosin-1), a peptide extracted from spider venom, was used to specifically target TP4 into cancer cells.
LYC1 with antimicrobial properties and 24 amino acid residues extracted from the venom of Lycosa singorensis demonstrated to have anticancer properties in vitro and in vivo in various cancers. By increasing Cyclin-Dependent Kinase (CDK) inhibitor proteins, LYC1 triggers apoptosis in cancer cell lines such as HeLa, A549, HT1080, HCT116, HepG2, etc. 15. The ability of this peptide to bind to cancer cell membrane to induce mitochondrial-dependent apoptosis has also been demonstrated 15. Furthermore, this peptide showed no toxic effects on normal cells such as HEK293 and erythrocytes. So, in addition to its specific cytotoxic effects, LYC1 can act as a suitable carrier for delivering of numerous cytotoxic agents to cancer cells as shown by Tan et al, in which LYC1 was used for the targeted transfer of gold nanoparticles to HeLa and SW480 cells 16.
The aim of this study was to synthesize a soluble recombinant fusion peptide, purify it using self-cleavage ability of inteins, and then assess its cytotoxic effects on the HeLa cell line, which serves as a model for cervical cancer cells.
Materials and Methods :
Bacterial strains, cell lines, plasmids and reagents: Top 10 and BL21 (DE3) Escherichia coli (E. coli) strains and cell lines including HeLa (Human cervical cancer cell line) and Human Umbilical Vein Endothelial Cell (HUVEC) were provided from Pasteur Institute of Iran (Tehran, Iran). Ampicillin and IPTG was purchased from Sigma (San Diego, California, USA). Chitin resin and pTWIN-1 vector were purchased from New England Biolabs (USA). The mentioned vector was transferred to Biomatik Company (Canada) for sub-cloning of desired genes including TP4-LYC1, TP4, and LYC1. All buffers for the purification were prepared according to the IMPACTTM manual, otherwise mentioned.
Soluble expression of TP4-LYC1, TP4, and LYC1 molecules: The selection of E. coli BL21 (DE3) colonies harboring the recombinant vectors including pTWIN-1-TP4-LYC1, pTWIN-1-TP4, and pTWIN-1-LYC1 was performed on LB-agar plates containing 100 µg/mL ampicillin. After the overnight cultivation, fresh cultures were inoculated and upon reaching an OD600 of 0.6, total expression of peptides in fusion to intein 1 and 2 of pTWIN-1 vector was induced by 1 mM IPTG for 4 hr at 37°C. Furthermore, in order to produce the mentioned proteins in soluble form, the expression was optimized at various IPTG concentrations (0.1, 0.3, and 0.5 mM) in different incubation temperatures as 7, 22, and 37°C for 17 hr. In each stage, the cells were harvested via centrifugation at 7000×g for 10 min at 4°C. Finally, evaluation of the protein expression was performed by 12% SDS-PAGE.
Peptide purification: The IMPACTTM purification system (New England Biolabs, Massachusetts, USA) was used to purify the recombinant peptides. Based on the N- and C- terminal cloning, the self-cleavage activity of inteins was induced by lowering the pH level from 8.5 to 6.5 for intein 1 and Dithiotritol addition to the related buffer for intein 2.
In detail, induction of intein cleavage from all fusion proteins was performed as follows: First, B1 buffer (Tris-HCl 20 mM, NaCl 500 mM, & EDTA 1 mM, pH=8.5) was added to the bacterial pellets and mixed using a vortex mixer. Then, for cell lysis, sonication was performed followed by the samples centrifuge at 7000 rpm for 30 min. The supernatant was poured into a column containing chitin resin and incubated for 30 min. The sample was passed through the column and then, the column was washed, at least 5 times, with B1 buffer. After that, 5 ml of B3 buffer (Tris-HCl 20 mM, NaCl 500 mM, EDTA 1 mM, 50 mM DTT, Triton-X100 0.3%, & Tween-20 0.2%, pH=8.5) was added to the column and left at room temperature for 24 hr. In the next day, the column supernatant was discharged and 5 ml of B2 buffer [Phosphate-Buffered Saline-(PBS), pH=6.5] was added to the column and put at room temperature for 24 hr. Finally, the purified column samples were collected and analyzed by Tris-Tricine SDS-PAGE.
Cytotoxic activity evaluation: The sterilization of each peptide was performed by 0.22 µM filtration and serial dilution of each peptide sample by the addition of PBS. Evaluation of the cytotoxic effects of recombinant peptides including TP4-LYC1, TP4, and LYC1 was performed by MTT assay on HeLa cancer cells compared with HUVEC as normal cells. Briefly, cells were cultured in RPMI 1640 culture medium enriched with 10% (v/v) Fetal Bovine Serum (FBS) and antibiotics (100 IU/ml penicillin and 100 µg/ml streptomycin) and incubated in the incubator at 37°C and 5% CO2. In this test, 180 µl of RPMI medium containing cells (in final concentration as 3×104 cell/ml) was added to each well of a 96-well plate and incubated at 37°C for 24 hr. The next day, 0.62, 1.25, 2.5, 5, and 10 µM of each peptide were added to the wells in the 20 μl final volume and incubated at 37°C for the other 48 hr. Exactly 20 μl of MTT solution (5 mg/ml) was added to each well and the plate was incubated in a CO2 incubator for 3 hr. Then, 150 μl of Dimethyl Sulfoxide (DMSO) was added to each well and slowly pipetted to dissolve the formazan crystals and the OD was measured at 570 nm using a microplate reader (Bio-Rad, US). PBS treated cells were used as the negative control and the cell free medium culture used as the blank.
Statistical analysis: In the biological assay stage, MTT test was repeated triplicate during independent experiments with four repeated wells for each concentration of peptides. The results were expressed as cell viability percent±SD. Data analysis was performed by SPSS 23 software. ANOVA and Tukey’s post hoc test was used to distinguish the differences between or among groups. The significance was assumed as p<0.05.
Results :
Expression and purification of peptides: SDS-PAGE analysis confirmed the expression of TP4-LYC1, TP4, LYC1 fused to intein 1 and 2 with bands at 60, 57, and 57 kDa, respectively. On the other hand, the optimum conditions for the soluble expression of each peptide was similar [0.5 mM IPTG at 22°C (Figures 1 and 2)]. After the induction of cleavage, the inteins 1 and 2 from the recombinant proteins, the appearance of bands at approximately 6, 3, and 3 kDa for TP4-LYC1, TP4, and LYC1, respectively on Tricine SDS-PAGE proved successful purification by the IMPACT system (Figure 3).
Finally, according to the Bradford method, in the optimized condition, the final yield of the purified peptides was calculated as 432, 571 and 477.5 μg/L of bacterial culture medium for TP4-LYC1, TP4, and LYC1, respectively.
Biological assay: The results of statistical analysis showed that approximately all peptides reveal their cytotoxic effects in a concentration-dependent manner. Actually, except LYC1 when used for the treatment of HUVEC cell line, all other peptides showed these effects. For HeLa cells, there was significant differences between the cytotoxic effects of the chimeric peptide in comparison to the negative control. Furthermore, the significant difference between the cytotoxicity of TP4-LYC1 and TP4 was established in the same concentrations. Finally, in the case of LYC1, the observed toxic effects were statistically lower than the two other peptide. However, a significant toxic effect was shown only in the higher concentration (10 µM) in comparison to the negative control (p-value=0.02) (Figure 4).
Regarding the HUVEC cells, the first finding is this fact that LYC1 showed no significant toxicity even in the higher concentration (10 µM) when compared to the negative control. Furthermore, in the same molar ratios there was no significant differences between the cytotoxicity of TP4 and TP4-LYC1. However, there was significant difference in the toxicity between TP4-LYC1 and the targeting moiety (LYC1) in the same concentrations (Figure 4).
Finally, based on the concentration vs. survival percent graph, the calculated IC50 of each surveyed peptides against two cell lines are reported as bellow:
For HeLa cells, the IC50 value was about 0.83, 2.75, and higher than 10 µM for TP4-LYC1, TP4 and LYC1. This value was not calculable for HUVEC cell line in the surveyed concentrations.
Discussion :
The aim of the present study was to produce a new recombinant fusion peptide containing TP4 with established cytotoxic effects and LYC1 as an AMP with safe profile against normal cell lines and successfulness in the usage as a drug delivery vehicle, in order to enhance the penetration ability of TP4 to cancer cells and diminish its toxic effects on the body normal cells and tissues.
In the first stage, the recombinant DNA technology was used as a suitable strategy to earn a fusion peptide containing 49 amino acid residues instead of its chemical synthesize in order to overcome the drawbacks of this method to access the enough amounts of this peptide. In the attempts to produce this chimeric peptide in soluble form and consequently with correct folding, we used intein tags and besides that, we used the optimization of the expression conditions in order to obtain the maximum amount of soluble peptides. The main concern in the production procedure of these AMP was their potency to kill the bacterial host cell after the production. We used several solutions to overcome this problem. First, we used lower post-induction temperature in order to diminish the activity of these antimicrobial peptides against E. coli expression system. Although lowering the temperature after the expression induction leads to diminish the protein expression, the chance of soluble expression increases. This strategy was used for the soluble expression of various peptides and proteins. For example, our previous fusion protein, DFF40-iRGD, was only produced at 7°C 17.
The other solution we used in this project was the fusion of the AMO to the intein tags, the large amino acid sequences in order to cover the alpha-helical structure of the peptides responsible for the bacterial membrane penetration and their antibacterial effects. In our pervious study, in an attempt to produce BR2 AMP, the fusion to the gyr A intein in its C-terminal was performed. Generally, production of various AMP in fusion to the inteins with auto-cleavage ability is a suitable approach that not only prevents their cell lysis activity, but also facilitates their purification stage. The other examples in this regard are human β-defensin 2 and LL-37 which are fused to the ΔI-CM mini-intein, expressed in aggregate form and after the self-cleavage induction, they were produced in soluble form with antimicrobial activities 18. In this example, it is obvious that in the expression stage, the produced AMP are safe for the expression of the host because of aggregation.
As shown in the results section, lowering the inducer concentration also leads to the soluble expression. Based on this fact, IMPACT protocol advises to use 0.4 mM IPTG as an inducer to improve the expression in soluble form. However, we used lower IPTG concentrations in order to diminish the cost of peptide production as well as improve the soluble expression. In our previous study, BIF1-iRGD fusion protein was only produced in soluble form by 0.05 mM of IPTG 19. In the present study 0.1 mM of IPTG leads to the soluble expression.
In the purification stage, on the other hand, we had to use two independent strategies in order to induce the auto-cleavage activity of two various inteins. The covering of two sites of peptides not only helps to prevent their bacterial lysis properties but also leads to the more soluble expression in comparison to the situations in which only one intein tag is used. However, the usage of two inteins complicates the self-cleavage induction. We have to use B2 buffer for inducing the intein 1 with pH=6.5 and B3 buffer containing DTT in order to induce the self-cleavage activity of intein 2. pH-shift in order to delete the intein 1 is a simple strategy for the cleavage of inteins but considering that the change in this item can easily occur at any stage of expression and purification, it leads to the induction of self-cleavage and reducing the final yield of peptide/protein product. One of the reasons for using low temperature in the expression stage is to reduce this process because the induction of self-cleavage occurs more likely at room temperature and the use of low temperatures in the expression stage prevents this process before the attachment of fusion protein to the chitin column. Various pH values were surveyed to determine the best one for more self-cleavage induction and it was shown that it is dependent to the protein type. For example, in our previous work for the recombinant production of a mutant form of IL-6R, the results showed that between three different pH, 4, 5.5, and 6.5, the most self-cleavage induction occurred at pH=4 20. While for the other protein, IL-1Ra, we produced more protein with a B2 buffer pH=6.5 21.
For the intein 2, on the other hand, the self-cleavage induction is a more complicated procedure than the intein 1; in this case, this process is related to the amino acid in the C-terminal of the target protein. Actually, for all produced peptides, (TP4 with an Arginine and TP4-LYC1 and LYC1 with a leucine at its C-terminal), down-stream intein (intein 2) was separated after 24 hr of incubation at room temperature. For BR2, terminated with an arginine residue, the same condition was used in our previous study 22.
Finally, in the case of biological study, as mentioned in the introduction section, TP4 was investigated for its anti-tumor effects in several studies. For example, Ting et al, surveyed the selective necrosis induction of this peptides against breast cancer cell lines including MDA-MB231, MDA-MB453, and MCF7 and their results showed that concentration of 7.5 µM of this peptide led to the significant toxicity after 24 hr of treatment in all cell lines especially MCF-7 and MDA-MB231 12. The concentration used in the mentioned study was similar to the concentrations in our study. However, we saw more toxicity of this peptide probably due to the more incubation time as 48 hr.
The other example in this regard is the investigation of antitumor activity of this peptide against A549 cancer cell. When this peptide was used in 18 µg/ml (6 µM) for 3 hr, microtubule network in this cell was disrupted. So, we can conclude that the concentrations which were surveyed in this study can be used for the other biological tests such as determination the cell death mechanism, microtubule disruption and so on. In the mentioned study, the penetration ability of this peptide was analyzed; TP4 showed penetration efficacy around the cancer cell membrane. However, there is no data about targeted penetration using normal cell lines 10. The other study in this regard, used normal lung cell lines as the control, and when compared to the normal cells, cancer cell lined showed more toxicity than normal cells but it seems that this difference is not significant based on their calculated IC50 for 3 hr incubation. However, over the time, this difference became more significant and this indicates that the cell entry of the peptide requires a minimum time. Another finding of this study was that this peptide activated the caspase-3 and induced apoptosis in 6.71 µM 11.
In our study, to further ensure the specific and targeted function of TP4, another peptide was added to its structure, which had been evaluated till now for the targeted delivery of gold nanoparticles to different cancer cells and showed no toxicity on normal cells such erythrocytes and HEK293 cells 16. It was the only study in this regard for targeting a therapeutic agent for cancer cells and as reported in the results section, LYC1 did not show any toxicity either on normal cells or on cancer cells. However, compared to TP4, the amount of toxicity against cancer cells increased in the chimeric peptide in equal molar ratios and this can confirm more penetration of the fusion protein than native TP4.
Conclusion :
In the present study, we tried to produce a recombinant form of chimeric peptide in order to decrease the chance of cytotoxic effects of TP4 against normal cells of the body. Although the production of these three peptides occurred in low yield, and also because of the suitable potential to act as a targeted therapy for the eradication of cancer cell, attempt to increase the final yield of production is extremely advised. In this regard, the usage of various auto-cleavage condition such as pH, different types of reducing agents, other additives such as Triton-×100 and tween-20 and finally changing the incubation time and temperature can be suggested for ongoing studies. However, other strategies for increasing the soluble protein production and purification are urgent stages in order to continue the other biological assays on the fusion peptide. Thus, this fusion peptide can act as a safe bio-molecule with potent cytotoxic effects against cancer cells. Meanwhile, in the first stage for the future studies in this regard, determination of penetration ability and the cell death mechanism must be performed in order to have more precise view in its action as a tumor targeted agent.
Acknowledgement :
The authors would like to appreciate valuable technical assistance of laboratory experts in molecular biotechnology and cell culture laboratories. The content of this paper was extracted from the grant with number as 198278, financially supported by Research Deputy of Isfahan University of Medical Sciences and approved by the ethics committee of IUMS with code IR.MUI. RESEARCH.REC.1398.820.
Funding: The content of this paper was extracted from the grant with number as 198278, financially supported by Research Deputy of Isfahan University of Medical Sciences.
Ethics Statement :
This article does not contain any studies with human participants or animals performed by any of the authors. The Ethics Committee of Isfahan University of Medical Sciences approved this research with the code of IR.MUI.RESEARCH.REC.1398.820.
Conflict of Interest :
All authors confirm that they have no conflict of interest.
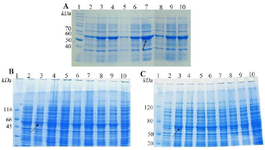
Figure 1. SDS-PAGE evaluation the total expression of E. coli BL21 (DE3) containing recombinant pTWIN1in various conditions. A) analysis of total LYC1 fusion protein expression. Lane 1: PageRuler™ unstained protein ladder 26614, Lane 2: induction by 0.1 mM IPTG at 7ºC, Lane 3: induction by 0.3 mM IPTG at 7ºC, Lane 4: induction by 0.5 mM IPTG at 7ºC, Lane 5: induction by 0.1 mM IPTG at 22ºC, Lane 6: induction by 0.3 mM IPTG at 22ºC. Lane 7: induction by 0.5 mM IPTG at 22ºC. Lane 8: induction by 0.1 mM IPTG at 37ºC. Lane 9: induction by 0.3 mM IPTG at 37ºC. Lane 10: induction by 0.5 mM IPTG at 37ºC. B). analysis of total TP4 fusion protein expression. Lane 1: PageRuler™ unstained protein ladder 26614, Lane 2: induction by 0.1 mM IPTG at 7ºC, Lane 3: induction by 0.3 mM IPTG at 7ºC, Lane 4: induction by 0.5 mM IPTG at 7ºC, Lane 5: induction by 0.1 mM IPTG at 22ºC, Lane 6: induction by 0.3 mM IPTG at 22ºC. Lane 7: induction by 0.5 mM IPTG at 22ºC. Lane 8: induction by 0.1 mM IPTG at 37ºC. Lane 9: induction by 0.3 mM IPTG at 37ºC. Lane 10: induction by 0.5 mM IPTG at 37ºC. C) analysis of total TP4 fusion protein expression. Lane 1: PageRuler™ unstained protein ladder 26614, Lane 2: induction by 0.1 mM IPTG at 7ºC, Lane 3: induction by 0.3 mM IPTG at 7ºC, Lane 4: induction by 0.5 mM IPTG at 7ºC, Lane 5: induction by 0.1 mM IPTG at 22ºC, Lane 6: induction by 0.3 mM IPTG at 22ºC. Lane 7: induction by 0.5 mM IPTG at 22ºC. Lane 8: induction by 0.1 mM IPTG at 37ºC. Lane 9: induction by 0.3 mM IPTG at 37ºC. Lane 10: induction by 0.5 mM IPTG at 37ºC.
|

Figure 2. SDS-PAGE evaluation the soluble expression of E. coli BL21 (DE3) containing recombinant pTWIN1in various conditions. A) analysis of total LYC1 fusion protein expression. Lane 1: PageRuler™ unstained protein ladder 26614, Lane 2: induction by 0.1 mM IPTG at 7ºC, Lane 3: induction by 0.3 mM IPTG at 7ºC, Lane 4: induction by 0.5 mM IPTG at 7ºC, Lane 5: induction by 0.1 mM IPTG at 22ºC, Lane 6: induction by 0.3 mM IPTG at 22ºC. Lane 7: induction by 0.5 mM IPTG at 22ºC. Lane 8: induction by 0.1 mM IPTG at 37ºC. Lane 9: induction by 0.3 mM IPTG at 37ºC. Lane 10: induction by 0.5 mM IPTG at 37ºC. B) analysis of total TP4 fusion protein expression. Lane 1: PageRuler™ unstained protein ladder 26614, Lane 2: induction by 0.1 mM IPTG at 7ºC, Lane 3: induction by 0.3 mM IPTG at 7ºC, Lane 4: induction by 0.5 mM IPTG at 7ºC, Lane 5: induction by 0.1 mM IPTG at 22ºC, Lane 6: induction by 0.3 mM IPTG at 22ºC. Lane 7: induction by 0.5 mM IPTG at 22ºC. Lane 8: induction by 0.1 mM IPTG at 37ºC. Lane 9: induction by 0.3 mM IPTG at 37ºC. Lane 10: induction by 0.5 mM IPTG at 37ºC. C) analysis of total TP4 fusion protein expression. Lane 1: PageRuler™ unstained protein ladder 26614, Lane 2: induction by 0.1 mM IPTG at 7ºC, Lane 3: induction by 0.3 mM IPTG at 7ºC, Lane 4: induction by 0.5 mM IPTG at 7ºC, Lane 5: induction by 0.1 mM IPTG at 22ºC, Lane 6: induction by 0.3 mM IPTG at 22ºC. Lane 7: induction by 0.5 mM IPTG at 22ºC. Lane 8: induction by 0.1 mM IPTG at 37ºC. Lane 9: induction by 0.3 mM IPTG at 37ºC. Lane 10: induction by 0.5 mM IPTG at 37ºC.
|
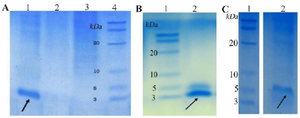
Figure 3. SDS-PAGE evaluation of purified peptides. A) purified LYC1. B) purified TP4. C) purified TP4-LYC1.
|
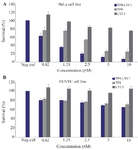
Figure 4. Cytotoxicity assays of LYC1, TP4, and TP4-LYC1 against HeLa (A) and HUVEC (B) cell lines. Error bars represent SD. stars were used to show the difference between TP4, and TP4-LYC1 anti-proliferative effects in the same concentrations.
|
|