One-step and Rapid Identification of SARS-CoV-2 using Real-Time Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP)
-
 Zeinoddini, Mehdi
Faculty of Chemistry and Chemical Engineering, Malek Ashtar University of Technology, Tehran, Iran, Tel: +98 21 22974604; Fax: +98 21 22962257; E-mail: zeinoddini52@mut.ac.ir
Zeinoddini, Mehdi
Faculty of Chemistry and Chemical Engineering, Malek Ashtar University of Technology, Tehran, Iran, Tel: +98 21 22974604; Fax: +98 21 22962257; E-mail: zeinoddini52@mut.ac.ir
-
 Fathi, Javad
Department of Bacteriology and Virology, School of Medicine, Shiraz University of Medical Sciences, Shiraz, Iran, Tel: +98 21 22974604; Fax: +98 21 22962257; E-mail: javadfathi70@yahoo.com
Fathi, Javad
Department of Bacteriology and Virology, School of Medicine, Shiraz University of Medical Sciences, Shiraz, Iran, Tel: +98 21 22974604; Fax: +98 21 22962257; E-mail: javadfathi70@yahoo.com
-
Keshavarz Alikhani, Hani
-
Department of Regenerative Medicine, Cell Science Research Center, Royan Institute for Stem Cell Biology and Technology, ACECR, Tehran, Iran
-
Shiekhi, Fatemeh
-
Faculty of Chemistry and Chemical Engineering, Malek Ashtar University of Technology, Tehran, Iran
Abstract: Background: SARS-CoV-2 as the cause of novel coronavirus disease (COVID-19) is a member of the family Coronaviridea that has generated an emerging global health concern. Controlling and preventing the spread of the disease requires a simple, portable, and rapid diagnostic method. Today, a standard method for detecting SARS-CoV-2 is quantitative real-time reverse transcription PCR, which is time-consuming and needs an advanced device. The aim of this study was to evaluate a faster and more cost-effective field-based testing method at the point of risk. We utilized a one-step RT-LAMP assay and developed, for the first time, a simple and rapid screening detection assay targeting the Envelope (E) gene, using specific primers.
Methods: For this, the total RNA was extracted from respiratory samples of COVID-19 infected patients and applied to one-step a RT-LAMP reaction. The LAMP products were visualized using green fluorescence (SYBR Green I). Sensitivity testing was conducted using different concentrations of the designed recombinant plasmid (TA-E) as positive control constructs. Additionally, selectivity testing was performed using the influenza H1N1 genome. Finally, the results were compared using with conventional real time RT-PCR.
Results: It was shown that the RT-LAMP assay has a sensitivity of approximately 15 ng for the E gene of SARS-CoV-2 when using extracted total RNA. Additionally, a sensitivity of 112 pg was achieved when using an artificially prepared TA-E plasmid. Accordingly, for the detection of SARS-CoV-2 infection, the RT-LAMP had high sensitivity and specificity and also could be an alternative method for real-time RT-PCR.
Conclusion: Overall, this method can be used as a portable, rapid, and easy method for detecting SARS-CoV-2 in the field and clinical laboratories.
Introduction :
On December 30, 2019, a new coronavirus emerged in Wuhan (Hubei Province, China) that caused the emergence of several pneumonia cases. It was similar to Severe Acute Respiratory Syndrome Coronaviruses (SARS-CoV). The World Health Organization (WHO) named this new virus SARS-CoV-2 based on its similar genome to SARS-CoV and the related disease was named as Coronavirus disease 2019 (COVID-19) 1-5. SARS-CoV-2 had a high potential for person-to-person transmission that was never found in humans, and eventually became an worldwide outbreak 1. Currently, SARS-CoV-2 has caused significant mortality (about 6 million deaths) and morbidity (about 300 million cases) in the world. SARS-CoV-2 is a member of the Betacoronavirus genus and genetically related to the Middle East Respiratory Syndrome coronavirus (MERS-CoV) and SARS-CoV 4,6. It was demonstrated that the genomic sequence of SARS-CoV-2 is closer to SARS-CoV than the MERS CoV. However, the amino acid sequence of SARS-CoV-2 was exclusively different from the other coronaviruses, especially, in the surface glycoprotein spike (S-protein) and the regions of ORF1ab polyprotein 1,7,8. Structurally, coronaviruses are 150 to 160 nm spherical nanoparticles containing five sections: positive single-stranded RNA (+ssRNA), nucleocapsid protein (N), membrane glycoprotein (M), envelope protein (E), and glycoprotein spike (S). More structural investigations showed that unlike SARS and MERS, the SARS-CoV-2 has an additional glycoprotein with acetyl esterase and hemagglutination (HE) properties 9,10.
The main method for detecting SARS-CoV-2 is the genome-based reverse transcription Polymerase Chain Reaction (RT-PCR). This method has a high speed, sensitivity and specificity as compared to conventional methods such as real-time PCR that have some limitations including, the use of thermocycler machine, high thermal cycles, time-consuming electrophoresis gel preparation, and urgent need for skilled personnel 11,12. Instead, the Loop-Mediated Isothermal Amplification (LAMP) isothermal technique can currently be used to identify SARS-CoV-2 in a rapid, easy, sensitive, and specific procedure. This technique was first introduced by Notomi in 2000 to detect the DNA of the hepatitis B virus 11. In the LAMP method, the heat-resistant Bst DNA polymerase with strand displacement activity was used that leads to high specificity in isothermal conditions. Another notable advantage of this technique is high sensitivity, specificity, and efficiency 13-17, the existence of various visual monitoring methods such as turbidity monitoring, and the use of fluorescent or colorimetric dyes without using an extensive tool such as thermocycler 16,17. In this study, we developed, for the first time, a one-step RT-LAMP assay for the detection of SARS-CoV-2 in infected patients using specific primers targeting the E gene. The results obtained from our assay were compared with those from Real-Time PCR.
Materials and Methods :
Patient samples: According to this experiments and observational study, total RNA was extracted from swabs obtained from 35 SARS-CoV-2 infected patients from the Diagnosis Laboratory of Chamran Hospital (Tehran, Iran) and was used for the validation of the one-step, one-tube, multiplex probe-based real-time PCR and RT-LAMP assay. The samples were anonymized without any patient’s specific characteristics including their age, gender, and clinic-pathological features.
Primer design: RT-LAMP specific primers were designed according to envelope (E) gene from the SARS-CoV-2 genomic sequence in GenBank (Accession No: MN908947) using PrimerExplorerV5 software (https://primerexplorer.jp) (Table 1). In this design, MEGA6 (Molecular Evolutionary Genetics Analysis Version 6.0) software was used for the multiple sequences alignment and the consensus sequences of E gene from SARS-CoV-2 were found (data not shown). Each set of primers contained two internal primers [Forward Inner Primer (FIP) and Backward Inner Primer (BIP)] and two external primers [Forward outer primer (F3) and Backward outer primer (B3)] (Table 1, Figure1).
Design of positive control constructs (TA-E): One intelligent strategy for designing genomic-based detection kits is the creation of positive control constructs from conserved regions of pathogen gene sequences. This approach ensures the availability of reliable positive controls that can be used to validate the performance of the detection kit, guaranteeing accurate and consistent results. This recombinant construct could be used also, for determining the sensitivity of the reaction. For this, the synthetic construct was designed using Snapgene offline software with the specific region of the envelope (E) fragment. Moreover, sequences retrieved from NCBI/EMBL were organized and aligned using the MEGA6 blast software (https://blast.ncbi.nim.nih.gov/Blast.cgi). In the next step, E segment was amplified using F3 and B3-specific primers. The PCR product was then cleaned-up and ligated into the TA-vector (pTG19-T PCR cloning Vivantis) and finally transformed into host cells (Escherichia coli DH5α). The produced recombinant bacteria were cultured in LB-broth media, and the extraction of the plasmid (named TA-E) was performed according to the GeneAll plasmid extraction kit (Korea). The extracted plasmids were quantified using a picodrop spectrophotometer and used as a template for PCR and LAMP amplification.
RNA extraction and RT-PCR assay: The total RNA was obtained from the clinical and respiratory samples taken from patients with COVID-19 at Chamran Hospital, extracted using RNX reagent according to the manufacturer’s instruction, and tested using the real-time RT-PCR method (Pishtaz Teb Diagnostics, Iran). For this, RT-PCR assay was performed in a total reaction volume of 10 µl containing 1 µl of 10×PCR buffer, 2 mM of MgSO4, 1.5 mM of dNTP, 1 mM of DTT, 1 µM of each of the B3 and F3 primers, 5 U of RNase inhibitor (Invitrogen), 5 U of cloned AMV reverse transcriptase (Invitrogen), 2.5 U of Taq DNA polymerase, 1.0 µg of the total RNA and RNase-free deionized water. The reactions were first incubated at 60°C for 60 min and then, the following steps were carried out: initial denaturation at 95°C for 5 min, 30 cycles of 95°C for 60 s, 60°C for 60 s, and 72°C for 60 s, and a final extension at 72°C for 10 min. The PCR reaction was done by an automated thermal cycler (Techgene, Germany). PCR products were electrophoresed using 1% agarose gel electrophoresis.
The one-step RT-LAMP detection: The RT-LAMP detection of SARS-CoV-2 was performed in a total reaction volume of 12.5 μl containing 1.25 μl of 10× isothermal amplification buffer, 4 mM of MgSO4, 1.4 mM dNTPs, 0.8 M of betaine, 0.1 μM of the outer primers F3 and B3, 0.8 μM of the inner primers FIP and BIP, 4 U of Bst 2.0 DNA polymerase (New England Biolabs), 5 U of cloned AMV reverse transcriptase (Invitrogen), 1.25 µl of DTT (0.1 M), 0.4 µl of RNasin and 60 ng of the total RNA. Finally, 1 µl of SYBR Green I (1/10 dilution in DMSO) was added into the microtube lid, and the mixture was incubated at 65°C for 60 min.
Specificity and sensitivity of RT-LAMP detection: The specificity and selectivity of RT-LAMP detection were determined by SARS-CoV-2 and Influenza H1N1. Also, to evaluate the sensitivity of RT-LAMP detection for SARS-CoV-2, concentrations of total RNA were reduced to 30 and 15 ng and also, the artificial construct (TA-E) was diluted to 112 pg, and then used as a template for RT-LAMP and LAMP detection, respectively.
Quantitative real time RT-PCR: The commercial COVID-19 One-Step RT-PCR kit (Pishtaz Teb Diagnostics, Iran) was used to detect the SARS-CoV-2 RNA. Briefly, each 20 µl reaction contained 5 µl buffer 49 (CAPITAL qRT-PCR Probe Mix), 0.5 µl of each primes, 0.25 µl probe, 1 µl enzyme RT 29, 2 µl extracted RNA and 8.5 µl RNase free dH2O. Rotor gene Q machine (Qiagen, Germany) was used for performing the reaction and the following program was set: reverse transcription at 50°C for 30 min, initial denaturation and enzyme inactivation at 95°C for 2 min, 45 cycles of extension at 95°C for 15 s and 60°C for 1 min for final extension, respectively 18.
Analysis of the RT-LAMP products: To visualize the RT-LAMP products, two methods were used: (1) after incubation at 60°C, SYBR Green that was placed into the microtube lid was mixed with the amplified product using quick spin mini centrifuge equipment, and the tubes were then visualized under a hand-held UV irradiation source and with eyes. (2) The amplicon was analyzed by running 5 µl of the amplified product on a 2% agarose gel and KBC power load staining (Kawsar Biotech, Iranian Company) for 60 min at 60 volts in 1×TAE (Tris Acetic acid EDTA) buffer and visualized under a gel documentation system. This test was performed in another laboratory outside the main lab.
Results :
Detection of SARS-CoV-2 by RT-PCR: RT-PCR detection was performed using F3/B3 primers. Amplification of 200 bp DNA (lane 1) fragments confirmed the SARS-CoV-2 genome in the total RNA extracted from the infected respiratory samples (Figure 2A). Also, after the TA-E plasmid production, the PCR assay was carried out and its product was analyzed using the agarose (1%) gel electrophoresis (Figure 2B).
Detection of SARS-CoV-2 by RT-LAMP: Evaluation of the RT-LAMP products was done by two methods; electrophoresis on 2% agarose gel and the addition of SYBR Green I after amplification. The results showed that DNA amplification was carried out using the total RNA (Figure 3A) and TA-E plasmid (Figure 3B) as a template compared to the negative control (without template).
Specificity and sensitivity of RT-LAMP: The selectivity of RT-LAMP assay for the detection of SARS-CoV-2 infection was guaranteed by 4 specific primers. Moreover, it was shown that the RT-LAMP method by the mentioned primers did not detect Influenza H1N1 (Figure 4A). In order to evaluate the sensitivity of RT-LAMP detections of SARS-CoV-2, we used different concentrations of the total RNA from 30 to 15 ng. Also, the sensitivity was determined using 11.25 ng to 112 pg of the TA-E plasmid. Data showed that the sensitivity of LAMP using total RNA and TA-E plasmid as targets were 15 ng and 112 pg, respectively (Figures 4B, 4C).
Evaluation and comparison of RT-LAMP with real-time PCR: The comparison of RT-LAMP assay with real-time PCR is shown in table 2. According to this table, among 35 samples, real-time PCR was able to identify 28 positive patients, out of 7 negative samples, 6 samples were without Cycle threshold (Ct), and these 6 samples were also detected as negative by RT-LAMP assay.
Discussion :
Some isothermal amplification techniques with straightforward features, low-cost, and accurate operations like LAMP techniques are suitable alternatives to conventional detection methods for infective agent screening 17,19-22. Today, it has been shown that the Limit of Detection (LOD) for the RT-LAMP technique in the detection of RNA viruses is approximately 10 copies per reaction which is tenfold more sensitive than RT-PCR 15. In 2020, Lambe developed a rapid method for the detection of COVID-19 using RT-LAMP assay, in which RT-LAMP specifically detected SARS-CoV-2 in less than 30 min 23. Moreover, Yu et al developed the iLACO (isothermal LAMP-based method for COVID-19) method for the amplification of a fragment in the ORF1ab gene. In this study, the species-specificity of iLACO was confirmed by comparing the sequences of 11 related viruses (including 9 coronaviruses and 2 influenza viruses) using the BLAST software. The sensitivity of this method was comparable to the Taq man-based real-time qPCR that could detect 10 RNA copies of SARS-CoV-2 24. Tholoth et al described a rapid, highly sensitive, and point-of-care molecular test that is amenable to be used by minimally trained individuals and using minimal equipment for SARS-CoV-2 detection in the clinical laboratory, at points of entry, and at home. This technique (named COVID-19 Penn-RAMP) is based on LAMP, in which two-step isothermal amplification was performed in order to increase the sensitivity to the nested nucleic acid. Both tests can be performed in closed tubes with either fluorescence or colorimetric dye and COVID-19 LAMP is carried out on par with COVID-19 RT-PCR. The results of this investigation showed that the sensitivity of COVID-19 RAMP is 10 to 100 folds better than COVID-19 LAMP and COVID-19 RT-PCR techniques 25.
Zhang et al also indicated a new method for RNA detection of SARS-CoV-2 from purified RNA or cell lysis using LAMP assay by colorimetric and visual detection. This assay was additionally confirmed using RNA samples purified from respiratory swabs collected from infected patients in Wuhan, with a similar function to an RT-qPCR commercial kit while requiring only heating and visual monitoring. This simple, rapid, selective, and sensitive assay provides an opportunity and capability to facilitate viral detection in the field without a requirement for complex diagnostic equipment. The results showed that all primer sets have similar detection sensitivity for the detection of lower than a few hundred copies, with sporadic detection of 120 copies. The results of the colorimetric detection method were similar to the real-time detection method described 26.
In the present work, we evaluated a real-time RT-LAMP detection for the envelope (E) gene of SARS-CoV-2 with high selectivity. In order to inhibit cross-contamination of LAMP products, we used a closed tube strategy by the addition of SYBR Green into the inner parts of microtubes. Also, the sensitivity of the method was determined using infected total RNA extracted from respiratory swabs, and the designed TA-E construct. Accordingly, data showed that the RT-LAMP sensitivity for envelope (E) gene of SARS-CoV-2 is about 15 ng and 112 pg from the extracted total RNA and artificially TA-E plasmid, respectively. In this study, the performance of RT-LAMP assay and real-time PCR was done for 35 infected samples. The comparison of these two assays showed that although the sensitivity of the RT-LAMP is lower compared to real-time PCR, more than 80% of viral RNA was detected, which can be ignored due to simplicity and lack of special equipment.
Conclusion :
The SARS-CoV-2 RT-LAMP detection developed in our study showed high sensitivity and selectivity. In conclusion, the one-step SARS-CoV-2 RT-LAMP directed for the envelope (E) gene was evaluated for the first time in this work that could be used in clinical samples.
Acknowledgement :
The authors would like to thank the research council of the Malek-Ashtar University of Technology (MUT) for the financial support of this investigation. This research was carried out with project number IR-MUT 299321801101, under the supervision of research management of Malek-Ashtar University of Technology.
Ethical approval :
The present study was performed on serum sample of suspected cases with SARS-CoV-2 infections at the Chamran Hospital, Tehran, Iran. Informed consent was obtained from all individual participants included in the study.
Conflict of Interest :
All authors have no competing interests in this research.

Figure 1. Locations of primer-binding sequences (Wuhan-Hu-1, com-plete genome, GenBank Accession No. MN908947.3).
|
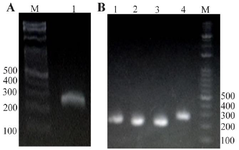
Figure 2. Analysis of RT-PCR product using 1% agarose gel electrophoresis. A) RT-PCR reaction with total RNA extraction from the patients' respiratory samples. Lane 1: RT-PCR product using primers F3/B3. B) PCR reaction with TA-E plasmid. Lane 1-4: PCR product using primers F3/B3, Lane M: 100 bp DNA leader.
|
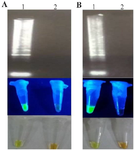
Figure 3. Analysis of the RT-LAMP reaction product with extracted total RNA (A) and TA-E plasmid (B) as a template. Lane 1: positive control, lane 2: negative control. Top: 1% agarose gel electrophoresis, middle: LAMP results indicated by SYBR green to the visualized under UV light, bottom: view the results with the naked eye.
|

Figure 4. A) The specificity of RT-LAMP reaction was determined using Influenza H1N1. Lane 1: total RNA extracted from SARS-CoV-2 infected respiratory samples, lane 2: RNA extracted from Influenza H1N1, lane 3: negative control. B) Sensitivity of RT-LAMP reaction using 30 (lane 1) and 15 ng (lane 2) from extracted total RNA of SARS-CoV-2 infected respiratory samples. C) Sensitivity of LAMP reaction using 11.25 ng (lane 1), 1.125 ng (lane 2), and 112 pg (lane 3) of TA-E plasmid. Top: agarose gel electrophoresis, middle: UV analysis, bottom: ocular observation.
|

Table 1. The specific primers for envelope (E) gene from SARS-CoV-2
|
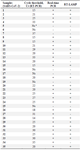
Table 2. Comparing the results of RT-LAMP method with real-time PCR on 35 patient samples
a: Negative sample
Cts <29 are strong positive, Cts of 30-35 are positive and Cts of ˃35 are negative.
|
|