Optimization of Degenerate PCR Conditions for Reducing Error Rates in Detection of PKS and NRPS Gene groups in Actinomycetes
-
Ghashghaei , Sara
-
Department of Cell and Molecular Biology & Microbiology, Faculty of Biological Science and Technology, University of Isfahan, Isfahan, Iran
-
 Etemadifar, Zahra
Department of Cell and Molecular Biology & Microbiology, Faculty of Biological Science and Technology, University of Isfahan, Isfahan, Iran, Tel: +98 31 37932367; Fax: +98 31 37932456; E-mail: z.etemadifar@sci.ui.ac.ir; zetemadifar@gmail.com
Etemadifar, Zahra
Department of Cell and Molecular Biology & Microbiology, Faculty of Biological Science and Technology, University of Isfahan, Isfahan, Iran, Tel: +98 31 37932367; Fax: +98 31 37932456; E-mail: z.etemadifar@sci.ui.ac.ir; zetemadifar@gmail.com
-
Tavassoli, Manoochehr
-
Department of Cell and Molecular Biology & Microbiology, Faculty of Biological Science and Technology, University of Isfahan, Isfahan, Isfahan, Iran
-
Mofid, Mohammad Reza
-
Department of Biochemistry, Isfahan Pharmaceutical Sciences Research Center and Bioinformatics Research Center, School of Pharmacy, Isfahan University of Medical Sciences, Isfahan, Iran
Abstract: Background: The screen of Polyketide Synthase (PKS) and Nonribosomal Peptide Synthetase (NRPS) gene groups is a quick way to discover new therapeutic agents. However, errors in laboratory techniques cause a loss of touch with reality. This study aimed to evaluate the presence of PKS and NRPS gene groups in previously isolated strains by optimizing their specialized amplification by degenerate primers and indicating the evolutionary relationships with reference strains.
Methods: PKS-I, II, and NRPS genes PCR amplification was performed using three degenerate primer sets for 22 actinomycete strains with antibacterial activity. Annealing temperature and the amount of template DNA and primers were optimized. PCR products of PKS-I, II, and NRPS from three strains were sequenced after TA cloning. Besides, strains with high antibacterial activity were identified by biochemical features and partial 16S rDNA sequencing and hypothetically classified by a phylogenetic tree.
Results: High frequencies of PKS-I (86.4%), PKS-II (81.8%), and NRPS (95.4%) genes were found among the strains after optimization. Fourteen strains (64%) contained all of the genes, and 100% of strains had at least two genes. These numbers are pretty distinct in comparison with the previous researches. All of the sequenced strains were members of Streptomyces genus.
Conclusion: Our research showed that degenerate PCR strongly depends on the variation of annealing temperature and primer concentration, resulting in an unexpected shift in PCR outputs. The sequencing results confirmed the optimized conditions for specialized PCR of PKS-I, PKS-II, and NRPS gene groups.
Introduction :
Actinobacteria represent a significant component of the microbial population in most soils. This phylum of bacteria has been extremely useful to the pharmaceutical industry due to its seemingly unlimited capacity to produce secondary metabolites with diverse biological activities and chemical structures. Approximately 50% of Actinobacteria are from the genus Streptomyces, and about 75% of commercially valuable antibiotics are derived from this genus 1.
However, novel technologies are demanded for discovering new bioactive secondary metabolites from these bacteria to meet the persistent and urgent medical need for newer and better drug candidates. Based on the hypothesis that secondary metabolites with similar structures are likely biosynthesized by gene clusters harboring certain homologous genes, such homologous genes could serve as appropriate selectable markers for different natural product biosynthetic gene clusters 2,3. A wide range of structurally diverse bioactive compounds, including toxins, siderophores, pigments, antibiotics, antitumor agents, and immunosuppressants, are synthesized on large Nonribosomal Peptide Synthetase (NRPS) and Polyketide Synthase (PKS) enzyme complexes 4-6. Therefore, screening for genes associated with secondary metabolism gives a better insight into the genetic capabilities of actinomycetes 2,3. NRPSs are composed of catalytic units called modules 6 working in an assembly-line fashion, where each module is responsible for incorporating a single monomer 7. According to the collinearity rule, the modules’ number and order define the number and order of amino acids in NRPs 6. Enzyme modules contain three core catalytic domains: adenylation (A), thiolation or peptidyl carrier (T or PCP), and condensation (C), respectively responsible for recognition and activation of an amino acid, transfer of the activated substrate to the peptidyl-carrier domain of the same module, and the formation of the C-N bond between the elongated chain and the activated amino acid 6,8. Actually, in the biosynthesis of NRPs, the enzymatic machinery itself functions as a template for the synthesized peptide product 7. PKs are synthesized through sequential decarboxylative condensation of the acyl CoA precursors by PKS. PKSs are classified into types I, II, and III 9. Type I PKSs, similar to type I Fatty Acid Synthase (FAS), are large multifunctional enzymes with a modular organization, with each module comprising catalytic domains 5,9,10. Typically, each module contains three essential catalytic domains for chain elongation: Ketosynthase (KS), Acyl Transferase (AT), and Acyl Carrier Protein (ACP). Bacterial type I PKSs are usually noniterative 11. Type II PKSs, similar to type II FAS, are multienzyme complexes acting in an iterative manner 9,11. They consist of a heterodimer formed by a KSα subunit (active site) and a KSβ or Chain Length Factor (CLF) subunit, which collaborate with ACP 12. Type III PKSs are homodimeric enzymes. Their single active site in each monomer catalyzes the priming, extension, and cyclization reactions iteratively to form PK products 9,11. Despite the structural simplicity, bacterial type III PKSs fall into five groups, many of which have been identified from Streptomyces 11. In addition to the common form of types I and II, exceptions such as iterative type I PKSs and non-iterative type II PKSs have also been reported by now 9. Due to the evolution of multidrug-resistant strains, screening of natural resources, especially actinomycetes, represents an important route to discover new therapeutic agents. In addition, the screening of PKS and NRPS genes is a quick way to show the potential for antibacterial agent production.
The overall aims of this study were (i) to evaluate the presence of PKS (types I and II) and NRPS gene groups involved in the biosynthesis of secondary metabolites in twenty-two antibacterial agent-producing actinomycete strains isolated in our previous study 13. For example, A3F and A7R primers target the conserved motifs A3 and A7 in adenylation domain. Therefore, PCR products can have different sizes (between 700-800) from the conserved motifs 3 to 7, (ii) to study the effect of different factors on degenerate PCR amplification, (iii) to scrutinize the power of the optimized reaction conditions for specialized amplification of PKS-I, PKS-II and NRPS gene groups by sequencing, and (iv) to show the phylogenetic and evolutionary relationships between some of the high potential antibacterial agent-producing strains and reference strains.
Materials and Methods :
Actinobacterial strains: Twenty-two antibacterial agent-producing actinomycete strains (F4, F6, F9, F10, Is1, Is4, Is5, Ih1, Ih6, Iz1, Iz2, Iz6, Iz8, Iz9, Iz11, Iz14, Iz16, Iz17, D2, B4, N1, and A2) isolated in our previous study 13 were selected for further investigation. All strains were scre-ened for the presence of PKS and NRPS genes. The high potential antibacterial agent-producing strains (F4, F6, F9, F10, Iz6, Iz8, Iz16, and B4) were also selected for molecular identification by 16S rDNA sequencing. The most active isolates against hospital-resistant strains and, in the next step, the strains with the largest halo diameter (Iz8, F9, and F6) were selected for cloning.
DNA extraction: DNA was extracted by CTAB method 14. The only difference for strain F9 was that a lysis treatment was applied at the beginning of protocol 15.
PCR detection of PKS-I, PKS-II, and NRPS genes: Genomic potential for producing bioactive metabolites was evaluated using three sets of specific degenerate primers: (1) K1F (5ˊ-TSAAGTCSAACATCGGB CA-3ˊ), and M6R (5ˊ-CGCAGGTTSCSGTACCAGT A-3ˊ) targeting PKS-I KS and methyl malonyl transferase domains 16-18; (2) KSαF (5ˊ-TSGCSTGCTTGG AYGCSATC-3ˊ), and KSαR (5ˊ-TGGAANCCGCCG AABCCTCT-3ˊ) targeting PKS-II KSα subunit 17,18; and (3) A3F (5ˊ-GCSTACSYSATSTACAC STCSGG-3ˊ), and A7R (5ˊ-SASGTCVCCSGTSCGGTAS-3ˊ) targeting the conserved motifs A3 and A7 in NRPS adenylation domain 16-18. PCR conditions for the initial amplification of PKS-I, PKS-II, and NRPS were as follows: a 25 µl sample of the PCR reaction mixture containing 10-300 ng of genomic DNA, 0.4-4 µM of each primer, 200 µM dNTPs, 1.5 mM MgCl2, 2.5 µl of 10x PCR buffer, 2.5 µl (1.4 M) of DMSO, and 2.5 U of Taq polymerase. The PCR was performed in an Eppendorf Mastercycler Gradient with the following cycling conditions: (i) 5 min at 95°C; (ii) 35 cycles of 30 s at 95°C, 2 min at 54-62°C (for K1F-M6R and KS𝛼F-KSαR) and 55-60°C (for A3F-A7R), and 4 min at 72°C; and (iii) 10 min at 72°C. Amplification products were analyzed by electrophoresis in 1% (w/v) agarose gels stained with ethidium bromide and bands of 1200-1400 bp, 600-700 bp, and 700-800 bp were classified as products of PKS-I, PKS-II, and NRPS genes, respectively 16-18.
TA cloning: PCR products of PKS-I, PKS-II, and NRPS associated with strains Iz8, F9, and F6, respectively, were selected to study the power of the optimized reaction conditions by sequencing. They were purified by gel purification kit (GeNet Bio, Korea) and cloned using the TA cloning kit (Thermo Fisher Scientific) as per the manufacturer’s instructions. Escherichia coli (E. coli) strain TOP10 was used as a cloning host. Then, colony PCR was used to confirm the length of the inserted product in all clones. The plasmid was extracted from one of the positive clones in each group by plasmid extraction kit (GeNet Bio, Korea) as per the manufacturer’s instructions. The purified plasmid DNAs were sequenced using universal primers M13F and M13R by Macrogen Inc. (Seoul, Republic of Korea). Sequences were tested against all DNA sequences available in GenBank database with the BLASTN program.
Identification and phylogenetic analysis of the high potential antibacterial agent-producing strains: Determination of cultural, morphological, and biochemical characteristics: The color of the sporulated aerial and substrate mycelia was examined on Starch Casein Agar (SCA) which had the following composition: 10.0 g/l soluble starch, 0.3 g/l casein, and basal mineral salt medium containing 2.0 g/l K2HPO4, 2.0 g/l KNO3, 2.0 g/l NaCl, 0.05 g/l MgSO4.7H2O, 0.02 g/l CaCO3, 0.01 g/l FeSO4.7H2O (scharlau, spain), and 15.0 g/l agar (Merck, Germany) 19,20. The pH of the medium was adjusted to 7.2 with the addition of HCl.
The utilization of different carbon sources (10 g/l) and degradation of starch (10 g/l) 21 and cellulose (2 g/l) 19 were studied in the basal mineral salt medium. The carbon sources were filtrated separately and added just before inoculation. Growth was compared with basal medium without carbon source as the negative control. The following carbon sources were tested: glucose, fructose, lactose, mannitol, raffinose, sorbitol, sucrose, and xylose. Proteolytic activity was determined using milk casein (plate) and gelatin (in tubes). Skimmed milk agar plates contained 5.0 g/l peptone, 3.0 g/l yeast extract, 1.0 g/l skimmed milk, and 12.0 g/l agar, pH=7.0 22. The gelatin hydrolysis test was performed on nutrient gelatin containing 100-150 g/l gelatin 23,24, final pH=7.2. Urease test media contained 1.0 g/l peptone, 1.0 g/l glucose, 5.0 g/l NaCl, 2.0 g/l KH2PO4, 0.012 g/l phenol red, 20.0 g/l urea, and 15.0 g/l agar. The pH of the medium was adjusted to 6.8 with the addition of HCl. The urea was filtered and added to the rest of the medium 25.
Molecular identification: The partial length sequence of 16S rDNA gene was amplified from the isolated genomic DNAs using the following two sets of universal bacterial 16S rDNA primers, "27F-YM (5′-AGAGTTTGATYMTGGCTC AG-3′) 26,27 and 1492R (5′-TACGGYTACCTTGTTA CGACTT-3′) 28" and "16SP0 (5′-GAAGAGTTTGATC CTGGCTCAG-3′) and 16SP6 (5′-CTACGGCTACCT TGTTACGA-3′)" 29. A 50 µl sample of the PCR reaction mixture contained 10-100 ng of genomic DNA, 0.4 µM of each primer, 200 µM dNTPs, 1.5 mM MgCl2, 5 µl of 10x PCR buffer, 5 µl (1.4 M) of DMSO, and 2.5 U of Taq polymerase, all obtained from CinnaGen Inc, Iran. The PCR was performed in an Eppendorf Thermal Cycler using appropriate programs optimized for each primer pair after denaturation of DNA by heating for 4 min at 94°C. The PCR program involved 35 cycles, and each cycle consisted of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, extension at 72°C for 2 min for "27F-YM/1492R" 28, and denaturation at 95°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 2 min for "16SP0/ 16SP6" 29. This was followed by a final elongation step for 5 min at 72°C. Moreover, we tested colony PCR without prior extraction of DNA as the template for rapid and simple detection of DNA sequences according to the method of Ishikava et al. 30 The PCR products were separated on a 1% agarose gel containing ethidium bromide in 1×TBE buffer. The purified PCR product was sequenced in both directions using an automated sequencer by Macrogen.
Phylogenetic analysis of 16S rDNA gene sequences: The sequences were edited using Finch TV V.1.4.0., then aligned with sequences from the closest related species of the genus Streptomyces retrieved from the GenBank/EzTaxon databases using PhyDE. The align-ment length was 1402 characters (including indels). Phylogenetic relationships were studied through Baye-sian inference using MrBayes version 3.2.6 31. JModeltest version 2.3 32 was used to find the best model of molecular evolution, and TIM1ef+I+G model was selected according to Akaike Information Criterion (AIC). Two parallel runs of four MCMC (Markov chain Monte Carlo) chains, including three heated chains and one cold chain, were carried out simultaneously for 8 million generations with a sample frequency of 1000. The analyses were stopped when the standard deviation of split frequencies was well below 0.01. The first 25% of generations were set to burn-in. Trees were visualized using TreeGraph2 33.
Results :
Study of the presence of PKS-I, PKS-II, and NRPS genes in twenty-two antibacterial agent-producing actinomycete strains: Twenty-two antibacterial agent-producing actinomycete strains (F4, F6, F9, F10, Is1, Is4, Is5, Ih1, Ih6, Iz1, Iz2, Iz6, Iz8, Iz9, Iz11, Iz14, Iz16, Iz17, D2, B4, N1, and A2) isolated in our previous study 13 (Table 1, Figure 1) were screened for the presence of PKSI, PKS-II and NRPS gene sequences by specific amplification of chromosomal DNA with primer sets K1F/ M6R, KS𝛼F/KS𝛼R, and A3F/A7R.
At first, multiple bands were found after PCR amplification of PKS-I genes with the annealing temperature estimated in previous researches 16-18 (Figure 2A only shows the results of some strains). Therefore, we tried to find the optimal annealing temperature for K1F/M6R primer set.
The optimal annealing temperature was variable for PKS-I depending on template DNA. It was: 55°C for Iz1, Iz11, Iz17, and Is1; 57°C for Iz2, Iz8, Iz9, Iz14, Is4, and Ih1; 58°C for B4 and D2; 59°C for F4, F9, and F10; 60°C for A2, Ih6, and N1, 61°C for F6 and Iz16; and 62°C for Iz6 and Is5. Concerning A2, all sub-bands were removed with the annealing temperature increased from 57 to 60°C. However, in some cases, we couldn’t get a single band at the mentioned temperatures, but as can be seen for Iz6, Iz16, F6, N1, and Ih6, the majority of non-specific bands with variable lengths were eliminated, and the rest were reduced to the minimum sharpness. (Figures 3A and 3B).
The optimal annealing temperature was 58°C for PKS-II because there was no shift from negative to positive at lower annealing temperatures, leading to non-specific DNA products (Figure 2B only shows the results of some strains). A comparison between Figures 2B, 3C and 3D shows that PCR amplification of F10, Iz6, and Iz8 at 58°C gives a single band on 600-700 bp while at 55°C gives multiple bands at different lengths. For NRPS genes, in addition to the specific band, a non-specific band was consistently observed at about 400-500 bp, and there was no change in the quality of the PCR output in the temperature range of 55-60°C. Some sequences were amplified at 55 up to 60°C, while some were amplified only at 55°C. Therefore, 55°C was the optimal annealing temperature for the degenerate primer set A3F/A7R (Figures 3E and 3F). Another factor that affects the success of PCR is primer concentration. The optimal concentrations to remove the false-negative results were 2, 0.8, and 2 µM for amplification of PKS-I, PKS-II, and NRPS genes, respectively. Actually, the results shifted from negative to positive or weak to strong by increasing primer concentrations from 0.8 to 2 µM for PKS-I and NRPS and from 0.4 to 0.8 µM for PKS-II. It is important to note that the number of non-specific bands sometimes increases with an excessive primer concentration. The minimum required amount of DNA that serves as the initial template or target sequence was 20-30 ng, and a higher amount did not have any marked effect on the outcome of PCR. Among investigated strains, 19 (86.4%), 18 (81.8%), and 21 (95.4%) strains were positive for PKS-I, PKS-II, and NRPS genes, respectively, after optimization (Table 2, Figure 3).
Sequence analysis of K1F/M6R, KSαF/KSαR, and A3F/ A7R PCR products: The clones selected from strains Iz8, F9, and F6 had high sequence similarity (85, 99, and 92%, respectively) with PKS-I gene on plasmid pSPA1 in Streptomyces parvulus (S. parvulus) strain 2297, the first PKSII gene previously reported in Streptomyces sp. strain MM1, and NRPS gene cluster in Streptomyces pactum (S. pactum) strain ACT12. These sequences were deposited in the NCBI GenBank database under the accession numbers KY637058, KY637060, and KY- 637057. The sequencing results confirmed the optimized conditions for specialized PCR of PKS-I, PKS-II, and NRPS gene groups.
Identification of morphological, biochemical, and molecular characteristics and assessment of phylogenetic relationships of eight actinomycete strains with high antimicrobial activity (F4, F6, F9, F10, Iz6, Iz8, Iz16, and B4): The ability of strains F4, F6, F9, F10, Iz6, Iz8, Iz16, and B4 to utilize various carbon sources for growth, proteolyze casein and gelatin, and degrade urea, was tested. All of them used glucose and starch, while none was able to use sorbitol and cellulose as the sole carbon source for growth. Every eight strains were urease-positive. "F4 and Iz8" and "F10 and Iz6" were able to degrade the same carbon sources (Table 3).
As shown in figure 4, amplification of 16S rDNA gene gives ≈1500 bp product using each of the two primer sets (six strains B4, F4, F6, F9, Iz6, and Iz16 using 27F-YM/1492R and two strains F10 and Iz8 using SP0/SP6). 16S rDNA gene of strains F10 and Iz8 had a very weak amplification by 27F-YM/1492R primer set, while, no product was observed by colony PCR.
Nearly full-length 16S rDNA gene sequences of all eight strains were compared with 16S rDNA sequences of type strains retrieved from GenBank/EzTaxon. All of them were assigned to genus Streptomyces and were deposited in the NCBI GenBank database under the accession numbers KX229767-73 (strains B4, F4, F6, F10, Iz6, Iz8 and Iz16, respectively) and KX417085 (strain F9) (Table 3).
Moreover, the alignment of 16S rDNA sequences showed a very high degree of similarity and conservation among the isolated strains. It was one of the most important reasons for using MrBayes instead of Mega to draw a phylogenetic tree. Phylogenetic analysis was performed by Bayesian inference implemented in Mr-Bayes based on the partially complete 16S rDNA gene sequences of eight selected strains and the type strains of closely related species (Figure 5). They were divided into four separate clades. Three strains of F9, F4, and B4 were placed in distinct clades of I, III, and IV, respectively, and five strains of F6, Iz16, Iz8, F10, and Iz6 were in a joint clade (clade II). Strain F9 and four strains, including Streptomyces chryseus (S. chryseus), Streptomyces helvaticus (S. helvaticus), S. albidocro-mogenes, and Streptomyces albosporeus (S. albospo-reus), formed a polytomic subclade within clade I, which was moderately supported (posterior probability=0.75). Clade II contained several differential sub-clades. Strain F6 was one of these subclades. Strains Iz8 and Iz16 are branched together on one sub-branch, while Iz6 and F10 are branched together on another with the posterior probability of 0.77 and 0.83. "Strain F4 and type strain of Streptomyces rectiviolaceus (S. rectiviolaceus)" and "strain B4 and type strain of Streptomyces tauricus (S. tauricus)" were in an internal subclade of clades III and IV, respectively, with the posterior probability>0.8.
Discussion :
NRPS and PKS pathways can be detected in many actinomycete isolates as the characteristics of secondary metabolite production 1. Prescreening of isolates with PCR primers targeting bioactive compounds-encoding genes (PKS-I, PKS-II, NRPS, etc.) is an effective and practical approach for detecting novel and useful secondary metabolites 34. On the other hand, the PCR detection of these genes is vital to discover isolates with high potential for the production of valuable secondary metabolites because some of the secondary metabolite biosynthetic gene clusters appear to be nonactive or silent under standard culture conditions, and they may require some specific triggers to be activated. From these, it can be concluded that the nonbioactive isolates hold their genetic capability for producing valuable secondary metabolites if they are cultivated under appropriate conditions 1. Therefore, it would be reasonable to utilize the screening of PKS and NRPS systems in combination with antimicrobial activity assay for biotechnological applications 18. Several research studies have been conducted and reported on the presence of PKS and NRPS genes as the characteristics of secondary metabolite production in Actinobacteria. It is noted that the low detection of PKS (types I and II) and NRPS gene fragments in the antibacterial agent-producing strains may reflect the lack of targeted genes or the implication of other genes in the biosynthesis of these bioactive products. Further, this may be caused by the types of selected degenerate primer pairs, which are unsuitable for amplifying these genes 2,5,18. Amos et al 35 validated two new updated PCR assays using new degenerate primers and concluded that these assays are specific, can detect clusters from a wide range of phyla, and have a good and reasonable hit rate.
To account for the discrepancies and inappropriate PCR conditions, twenty-two strains were screened for the presence of PKSI, PKS-II, and NRPS gene sequences by specific amplification of chromosomal DNA with primer sets K1F/M6R, KS𝛼F/KS𝛼R, and A3F/A7R 16-18. Actually, in the previous researches, PCR was performed at the fixed annealing temperature of 67.5°C 36 for A3F/A7R, 56 3 or 57°C 1,2,17,18,34 for K1F/M6R and A3F/A7R and 57 1, 58°C 2,17,18,34 or 63°C 3 for KSαF/KSαR and KSαF/KSβR. Lee et al 1 used the annealing temperature of 62°C for A3F/A7R. In contrast, our analysis showed that PCR amplification of the KS domain belonging to PKS-I using the degenerate primer set K1F/M6R needs different annealing temperatures depending on the template DNA. This is not inconsistent with the fact that some strains do not have this gene but may raise the possibility of getting the actual positive results. Moreover, some sequences can amplify only at a lower annealing temperature (55°C) by primer set A3F/A7R.
Another factor that affects the success of PCR is primer concentration, as sometimes the low concentration might lead to an undetectable product giving a false negative result. The optimal concentrations were 2 µM for PKS-I and NRPS and 0.8 µM for PKS-II, while previous researchers used primers at the final concentration of 0.2-0.8 µM 1-3,17,18,35. The dependence of amplification efficiency on the template amount was low. In this study, 14 strains (64%) contained all of the studied genes, and 100% of strains had at least two of the target genes. These numbers are quite distinct in comparison with the previous researches: 19.5, 42.5, and 5.7 1; 34.1, 63.4, and 61 2; 10.9, 15.2, and 26.1 34; 11.5, 23, and 76.9 17; 46.6, 60, and 46.6 18; 79.6, 70.4, and 57.4; and 65.6, 45.9, and 34.4 3 for PKS-I, PKS-II and NRPS genes, respectively.
The sequencing results indicated the high presence of genus Streptomyces compared to other actinomycetes in our soil samples. However, it should be noted that starch casein agar, containing starch as the sole carbon source, was the only medium used to separate these actinomycete strains 13. It is well accepted that members of the genus Streptomyces are prolific producers of novel antibiotics, with approximately 75% of commercially valuable antibiotics being derived from Streptomyces 1.
Conclusion :
The screening of PKS and NRPS genes is a quick way to show the potential for antimicrobial agent production. However, errors in laboratory techniques sometimes can make us move away from reality. The optimization of degenerate PCR will help us reduce error rates in the lab. Based on the results obtained in the present study, it can be concluded that degenerate PCR strongly depends on the annealing temperature and primer concentration, as any changes in these parameters can result in an unexpected shift in PCR outputs. Actually in degenerate PCR, we are dealing with different primers, and naturally, they have different annealing temperatures. For example, the primer set KSαF/ KSαR can be combined in 192 modes, and the annealing temperature will be different depending on which F and R primers are matched with DNA. Moreover, in some cases, bacteria in one common clade produce identical antibiotics. The phylogenetic tree helps us to choose different antibiotic-producing strains and avoid the purification of similar antibiotics by classifying the strains.
Acknowledgement :
This work was supported by the University of Isfahan, Iran. We would like to express our deepest appreciation to Maryam Malekmohammadi from University of Tehran for her assistance.
Conflict of Interest :
The authors declare that they have no conflict of interest.

Figure 1. The antimicrobial activity of several actinomycete strains against hospital-acquired bacterial and fungal isolates.
|
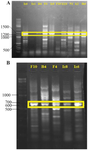
Figure 2. PCR amplification of PKS-I (A) and PKS-II (B) gene sequences with an amplicon size of 1200-1400 and 600-700 bp, respectively, for a number of strains at 57 and 55ºC, respectively. The name of strains has mentioned at the top of gel.
|
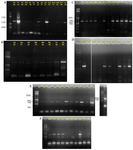
Figure 3. PCR amplification of PKS-I (A and B), PKS-II (C and D) and NRPS (E and F) gene sequences with an amplicon size of 1200-1400, 600-700 and 700-800 bp using primers K1F/M6R, KSαF/KSαR and A3F/A7R, respectively, on agarose gel 1%. The name of strains has mentioned at the top of gel.
|
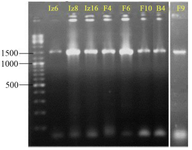
Figure 4. Agarose gel electrophoresis of 16S rDNA gene PCR-amplified region. The ladder includes fragments ranging from 100-10,000 bp. The name of strains has mentioned at the top of gel.
|

Figure 5. Bayesian inference tree showing the phylogenetic relationship among the eight selected strains and the type strain species of Streptomyces with the highest 16S rDNA sequence similarity. Posterior probabilities for the nodes are shown under the branches.
|
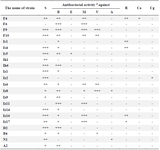
Table 1. Antibacterial activity of some actinomycete strains isolated from soil samples of Iran
a: Estimated by measuring the mean diameter of the clear zone of growth inhibition obtained from triplicate. Symbols: S, Staphylococcus aureus (ATCC 6538); B) Bacillus subtilis (ATCC 12711); E) Escherichia coli (ATCC 1399); M) methicillin-resistant Staphylococcus aureus; V) vancomycin-resistant enterococci; A) Aspergillus fumigatus; R, Rhizopus sp.; Ca, C. albicans; Cg, C. glabrata; -, no activity; +, weak activity; ++, moderate activity and +++, strong activity.
|
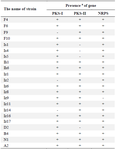
Table 2. The presence of PKS-I, PKS-II and NRPS gene groups in twenty-two antimicrobial agent-producing Streptomyces strains
a: +, present; -, absent.
|
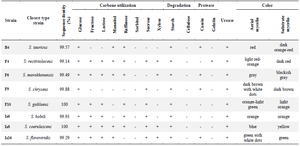
Table 3. Morphological, physiological and biochemical characteristics and molecular identification of the high potential antibacterial agent-producing strains
+, positive result; -, negative result
|
|