Identification of Critical Molecular Factors and Side Effects Underlying the Response to Thalicthuberine in Prostate Cancer: A Systems Biology Approach
-
Saberi , Fatemeh
-
Student Research Committee, Department of Medical Biotechnology, School of Advanced Technologies in Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
-
Cellular and Molecular Biology Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
-
Dehghan , Zeinab
-
Department of Comparative Biomedical Sciences, School of Advanced Medical Sciences and Technologies, Shiraz University of Medical Sciences, Shiraz, Iran
-
Noori , Effat
-
Student Research Committee, Department of Medical Biotechnology, School of Advanced Technologies in Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
-
Cellular and Molecular Biology Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
-
Taheri, Zahra
-
Department of Biology and Biotechnology, Pavia University, Pavia, Italy
-
Sameni, Marzieh
-
Student Research Committee, Department of Medical Biotechnology, School of Advanced Technologies in Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
-
Cellular and Molecular Biology Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
-
 Zali, Hakimeh
Department of Tissue Engineering and Applied Cell Sciences, School of Advanced Technologies in Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran, Tel: +98 21 22439848; E-mail: Hakimehzali@gmail.com
Zali, Hakimeh
Department of Tissue Engineering and Applied Cell Sciences, School of Advanced Technologies in Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran, Tel: +98 21 22439848; E-mail: Hakimehzali@gmail.com
Abstract: Background: Uncontrolled mitosis of cancer cells and resistance cells to chemotherapy drugs are the challenges of prostate cancer. Thalicthuberine causes a mitotic arrest and a reduction of the effects of drug resistance, resulting in cell death. In this study, we applied bioinformatics and computational biology methods to identify functional pathways and side effects in response to Thalicthuberine in prostate cancer patients.
Methods: Microarray data were retrieved from Gene Expression Omnibus (GEO), and protein-protein interactions and gene regulatory networks were constructed, using the Cytoscape software. The critical genes and molecular mechanisms in response to Thalicthuberine and its side effects were identified, using the Cytoscape software and WebGestalt server, respectively. Finally, GEPIA2 was used to predict the relationship between critical genes and prostate cancer.
Results: The POLQ, EGR1, CDKN1A, FOS, MDM2, CDC20, CCNB1, and CCNB2 were identified as critical genes in response to this drug. The functional mechanisms of Thalicthuberine include a response to oxygen levels, toxic substances and immobilization stress, cell cycle regulation, regeneration, the p53 signaling pathway, the action of the parathyroid hormone, and the FoxO signaling pathway. Besides, the drug has side effects including muscle cramping, abdominal pains, paresthesia, and metabolic diseases.
Conclusion: Our model suggested newly predicted crucial genes, molecular mechanisms, and possible side effects of this drug. However, further studies are required.
Introduction :
Prostate Cancer (PC) is neoplastic disease with significant clinical and economic burden that affects millions of men worldwide, especially in developed countries 1,2. PC is the second most common cause of substantial morbidity and mortality in men. PC prevalence is estimated to be approximately 23% of all male cancers in 2012 in the population of Europe. This rate is increasing significantly over recent years. The disease is complex and morphologically heterogeneous but is classified as adenocarcinoma 1. PC generally has 99% overall survival for ten years if detected and treated early 3. However metastatic castrate-resistant PCa (mCRPC) form remains incurable 2,4.
The course of the disease is influenced by factors such as the individual's health condition and the histopathological, anatomical, and molecular profile of the tumor. PC can be considered an age-related disease because its prevalence increases exponentially with age 5. Given the aging population of the world, this could be an alarm. In addition, environmental factors associated with PC include radiation exposure, a high intake of saturated fat, dietary deficiency of selenium, vitamin E, and D. The overexpression of mutant Tp53 (Tumor protein) and B-cell lymphoma 2 (bcl-2), as well as reinforcement of the androgen receptor, play key roles in the development of the refractory hormone phenotype 6,7. Also, Mutated Mutated in Multiple Advanced Cancers 1/P10 (MMAC1/p10) tumor suppressor genes may play a role in the metastatic phenotype of PC. Impotence and impaired urinary and bowel function are long-term side effects of over-treatment and treatment PC 1.
The feature of cancer cells is that they divide uncontrollably at a high proliferation rate. Mitosis plays a fundamental role in cancer progression and is one of the most effective targets of cancer chemotherapy 6. Volasertib and Alisertib (MNL8237), inhibitors of vital regulators of mitosis, have shown hopeful clinical efficacy in a variety of malignancies, including PC and acute myeloid leukemia. To date, microtubule-targeting drugs are the most effective mitosis inhibitors in chemotherapy strategies 4. Docetaxel and cabazitaxel are widely used to treat PC in a chemotherapeutic approach. However, severe toxic effects and the development of drug resistance are among the concerns of using these drugs 8. Therefore, action investigation of new therapeutic agents such as Thalicthuberine (TH) is an unmet clinical need. TH is a phenanthrene alkaloid isolated from various sources. This natural product induces cell death in malignant human cell lines through apoptosis. Studies have shown that TH indirectly targets microtubule dynamics by inhibiting a tubulin-associated protein. One of the unresolved challenges in the treatment of cancer is drug resistance. P-glycoprotein (Pgp) is responsible for multidrug resistance in many cancers 4. TH is not the primary substrate for Pgp and can treat drug-resistant cancers. This issue highlights the importance of studying the precise molecular mechanisms of this substance. Despite the improvement and survival of patients, anticancer drugs have various side effects such as damaging normal tissue and interrupting cell division of normal cells, which is a source of concern for patients and clinicians. There is a slight difference between an effective dose and a toxic dose of anticancer drugs. Therefore, sufficient knowledge of the side effects of anticancer drugs will help us to use the appropriate dose of anticancer drugs and manage unfavorable side effects observed during the therapy 9.
Recently, bioinformatics approaches and network-based methods have allowed us to find out the precise molecular mechanisms of drugs and probably their side effects. A deep understanding of the functional mechanisms of cancer is one of the essential components in the personalized and lifelong management of such diseases. Furthermore, these new biology concepts would help develop efficient and novel therapeutic and preventive approaches 10-12. Utilizing and analyzing Gene Regulatory Networks (GRNs) could get insights into the outcomes of clinical trials 13. In recent years' GRNs, high-throughput technologies, have been used for the analysis of gene expression data 14. GRNs are a collection of molecular regulators that interact with each other and regulate the gene expression levels of mRNA and proteins. Gene regulatory networks play a key role in cellular function 15. Motifs in GRNs have an important function in determining gene expression such as generating temporal expression programs and governing the responses to fluctuating external signals 16. Omics technologies led to novel classifications of prostate tumors and helped to choose novel targeted therapies 17.
The detailed molecular mechanisms of TH in PC treatment and probably its side effects are not still comprehensively understood. Therefore, this study re-ports Protein-Protein Interaction (PPI) and gene regulatory networks to identify essential functional mechanisms and biological functions in response to TH. Here, we employ the Protein-Protein Interaction Network (PPIN) and GRN to predict and identify drug targets, Gene Ontology's (GOs), and PC's biochemical pathways in response to TH and probable side effects.
Materials and Methods :
This study is divided into four steps: 1) the identification of appropriate Genomic Spatial Event (GSE) of microarray data, including PC (LNCap cell line)/TH and non-treatable PC (LNCaP cell line) samples, 2) the analysis of PPIN and GRN of a dataset, 3) the identification and enrichment analysis of crucial genes and finally 4) the prediction of probable side effects. Figure 1 describes the workflow of the study.
Data collection and processing: The PC/TH dataset was extracted from the Gene Expression Omnibus (GEO) database (http://www.ncbi. nlm.nih.gov/geo/) 18. The GSE83459 dataset was collected as a dataset comparing PC (LNCaP cell line)/TH and non-treatable PC (LNCaP cell line) 4. Differentially-Expressed Genes (DEGs) were identified according to (p<0.05) and (log2 fold change >0.5 and <-0.5. These DEGs were used for further analysis.
PPIN construction and topological analysis: DEGs were plugged into the Bisogenet app (HPRD database) and STRING (https://string-db.org/) separately, and the interactions with threshold 0.7 were selected to retrieve the protein interaction network. We merged two networks in Cytoscape 3.5.1 software 19 and investigated the network topologically. Network Analyzer was used for the identification of nodes with high degree (hub), Betweenness-Centrality (bottleneck), and Closeness-Centrality. The nodes with degree (hub) >1 were used as functional critical nodes. Both hub and bottleneck with high Closeness-Cen-trality play essential roles in fundamental cellular processes 20,21.
Construction of PPI subnetwork: The Molecular Complex Detection (MCODE) app was applied to screen sub-network with highly interconnected regions in the protein network and identified their seed genes. The MCODE parameters included Node Score Cutoff: 0.2, Degree Cutoff: 2, K-Core: 2, and Max-Depth: 100 22,23. The sub-networks with highly interconnected regions and their seed genes are the highest degree value in a network 22,24
Gene regulatory network: The identification of miRNAs suppressing DEGs: We obtained experimentally validated miRNA-target gene interactions from miRTarBase release 8.0 (http://miR TarBase.mbc.nctu.edu.tw/) and miRecords (http://c1. accurascience.com/miRecords/). The miRTarBase is a curated database from experimentally validated miRNA-target gene interactions and includes more than 13,404 validated interactions 25. The miRrecords is an experimental database of validated miRNA-target interaction 26.
The identification of transcription factors (TFs) regulating DEGs: We extracted TFs regulating DEGs from TRANSFACT (http://genexplain.com/transfac/) and TRRUST v2 (www.grnpedia.org/trrust/) databases. TRANSFACT is a curated database of eukaryotic transcription factors 27. TRRUST is validated experimental data from 800 and 828 TFs for humans and mice, respectively 28.
The identification of miRNAs suppressing TFs: We obtained miRNAs regulation TFs from miRTar- Base and miRecords databases.
The identification of TFs regulating miRs: We extracted TFs regulating miRNAs of TransmiR v2.0 (http://www.cuilab.cn/transmir). This database is a curated data of 3730 TF-miRNA regulations related to 19 species from >8000 publications 29.
GRN construction and motif detection: The network motifs are highly significant patterns that include molecular interactions and biological functions 30,31. To identify the regulatory motifs, we obtained regulatory relationships (TF-miRNAs, TF-Gene, miRNA-Gene, and miRNA-TF). We then merged and fed relationships into FANMOD software for obtained motifs with three nodes. FANMOD is a tool for detecting motifs in a big network and colored motifs analysis. This tool uses the RAND-ESU novel algorithm 32. The 3-node motifs were generated randomly 1000 times and compared with the input network. The 3-node motifs with Z-score >2.0 and p<0.05 were selected as significant motifs 33. The motifs with the same ID were merged and created a unique motif. Four relationships were merged in Cytoscape 3.5.1 software and motifs were extracted from GRNR. Finally, 10% of nodes with a high degree of centrality and closeness centrality were identified in the new network. The intersection of 10% of nodes with high degree, betweenness centrality, and closeness centrality was identified as the final network for further analysis.
Functional analysis: We performed enrichment analysis for Gene Ontology (biological process, molecular function, and cellular component) and Kyoto Encyclopedia of Genes and Genomes (KEGG) on DEGs using WebGestalt (http:// www.webgestalt.org) 34. The gene ontology and KEGG pathways with p<0.05 were identified as statistically significant terms. Finally, the key genes, biological processes, and functional pathways were discussed and verified by the experimental studies available in the literature review to identify active mechanisms and probable side effects related to this drug.
Confirmation of critical genes: The Gene Expression Profiling Interactive Analysis 2 (GEPIA2) is a web server for interactive analysis including, differential expression analysis, profiling plotting, correlation analysis, patient survival analysis, similar gene detection, and dimensionality reduction analysis 35. This web resource analyzes RNA sequencing data using a standard processing pipeline based on GTEx (Genotype-Tissue Expression) and TCGA (The Cancer Genome Atlas) (http://gepia.cancer-pku.cn/). We used GEPIA2 to evaluate the relationship between our critical genes and cancer development by Kaplan-Meier and boxplots.
Results :
Change in gene expression related to PC (LNCaP cell line)/TH: This study first investigated how DEGs and their related TF/miRs regulate gene ontology and pathways related to PC in response to TH, using PPIN and GRN. We identified the DEGs based on p<0.05 and log2 fold change >0.5 and <-0.5. The obtained DEGs include 8 down-regulated genes and 30 up-regulated genes used for further analysis. Supplementary table S1 reported these DEGs.
PPIN construction and topological analysis: We obtained the PPIN using the Bisogenet app (HPRD) and STRING databases. These networks merged to create a PPIN. The PPIN was visualized and topologically analyzed by Cytoscape software. This PPIN had 38 nodes and 79 edges. The propertied network included a clustering coefficient of 0.301, network diameter of 6, network density of 0.077, and shortest path of 466 (33%). The DEGs with degree>1 were investigated as critical genes. The list of these genes with a degree >1 is represented in figure 2A and table 1. The PPI clusters are highly connected regions of the network. The MCODE app identified these sub-networks in Cytoscape software. One sub-network with seven nodes, 24 edges, and a score of 6.667 was identified as a cluster with highly connected regions in the PPIN. The PLOQ gene was identified as a seed node in this cluster (Figure 2B).
Gene regulatory network construction and module detection: We collected four regulatory relationships (miR-gene, miR-TF, TF-gene, and TF-miR) for DEGs (Table 2, Supplementary Table S2). We imported and merged the four regulatory relationships in Cytoscape software as GRN. The four relationships fed into FANMOD software and 3-node motifs of GRN with at least two different color edges and p<0.05 and Z-score >2 were selected as motifs for further analysis (Figure 3).
We then merged the motif-related sub-networks with the same FAN MOD ID and extracted them from GRN. Finally, shared genes of 10% betweenness centrality, closeness centrality, and genes with the highest degree were selected as crucial genes. Figure 4 depicts essential genes.
Gene ontology and functional pathways enrichment analysis: The enrichment analysis on the DEGs (38 genes) was performed using Webgestalt (p<0.05). The biological processes included response to immobilization stress, response to oxygen levels, regulation of cell cycle, and response to a toxic substance. The molecular function term of DEGs was cyclin-dependent protein kinase activity, and most of the DEGs were presented in the spindle. The significant KEGG pathways were the p53 signaling pathway, cell cycle, parathyroid hormone synthesis, secretion and action, and FoxO signaling pathway. These results are represented in table 3. Finally, we investigated the role of crucial genes and biological processes, and KEGG pathways in probable side effects created by this drug using a literature review.
Confirmation of POLQ, EGR1, CDKN1A, FOS, MDM2, CDC20, CCNB1, and CCNB2 critical genes using GEPIA2: The network analysis results showed POLQ, EGR1, CDKN1A, FOS, MDM2, CDC20, CCNB1, and CCNB2 as critical genes in Levrier C et al study, in which a human PC cell line was treated with TH. In this study, Kaplan-Meier and box plots were used to demonstrate critical genes of network analysis. Using boxplot, we demonstrated the expression level of critical genes in PC, a significant over-expression of CDC20 and CCNB1 in PC in comparison with normal tissue, and a non-significant expression level of POLQ, EGR1, CDKN1A, FOS, MDM2, and CCNB2 genes (Figure 5). Figure 6 compares the overall survival rate of PC patients due to the high and low expression of POLQ, EGR1, CDKN1A, FOS, MDM2, CDC20, CCNB1, and CCNB2 genes. The results determined that among critical genes the decrease in CDKN1A and MDM2 expression harmed the survival of the patients. Besides, over-expression of EGR1, FOS, and CCNB1 also reduced the survival ability of the patients.
Discussion :
PC is the second most common cancer in developed countries and the fifth most common cause of cancer death. One of the challenges of treating this disease is cell resistance to chemotherapy drugs. Uncontrolled mitosis is the cancer cell mechanism, requiring the development of microtubule-targeting drugs. TH causes cell death by arresting mitosis and also reduces the effects of drug resistance. TH is a natural product extracted from the Australian plant Hernandia Albiflora Kubitzki. This novel anti-mitotic agent in prostate and cervical cancer cells escapes from the resistance mechanisms 4. There is still no comprehensive data on the functional mechanisms of TH in the treatment of PC. For further experimental examinations, systems biology and bioinformatics studies can help to identify critical genes and mechanisms mediating PC in response to TH.
Recently, network-based approaches have prepared powerful tools to identify the molecular function and complex processes of diseases by regulatory and MCODE clusters in GRN and PPIN. The nodes participating in these specific regions usually have key roles in cellular functions 36. The present study is the first in silico analysis to predict critical genes and molecular mechanisms of PCs treated with TH and its side effects. In this study, eight genes as critical nodes were introduced to PPIN and GRN in the LNCaP cell line in response to TH. Our analysis of PPIN and GRN of the LNCaP cell line in response to TH demonstrated that POLQ, EGR1, CDKN1A, FOS, CDC20, MDM2, CCNB1, and CCNB2 are critical genes. The functional enrichment analysis indicated that TH is involved in response to oxygen levels, immobilization stress and toxic substance, cell cycle regulation, regeneration, p53 signaling pathway, parathyroid hormone synthesis, secretion, and action, and FoxO signaling pathway.
POLQ, DNA polymerase θ, a versatile enzyme involved in DNA repair pathways, will be up-regulated in the PC cell line in response to TH. This gene was identified as a seed gene in the MCODE cluster of PPIN. This factor is involved in especially error-prone double-stranded fractures. DNA repair genes are often mutated in cancer cells and control of these genes will be a targeted therapeutic strategy expected from this drug 37. The study of Whole-exome sequencing of Indian PC by Febina Ravindran et al in 2022 showed POLQ can be introduced as the druggable target in the treatment of PC 38.
EGR1, CDKN1A, CCNB1, CCNB2, MDM2, and FOS were identified as hub-bottleneck in PPIN and GRN. EGR1 is an essential factor that will be down-regulated in response to TH in the PC cell line. This gene is an oncogenic transcription factor associated with PC. EGR1 regulates tumor suppressors such as PTEN, P53, TGFβ1, and fibronectin. PTEN and P53 are inactive in a majority of cancers. Defects in these suppressor genes are due to deregulated EGR1 39. Lechen Li showed that EGR1 can promote metastasis and regulate angiogenesis and osteoclastogenesis in PC 40. Therefore, down-regulation of EGR1 in PC treated with TH could help to treat patients.
P21 (CDKN1A), a Cyclin-Dependent Kinase Inhibitor (CDKI), is one of the up-regulated genes involved in the mechanism of this drug. This gene plays a pivotal role in cell cycle development regulating CDK activity. P21 is known for its dual function. On the one hand, it acts as a tumor suppressor gene that induces cell cycle arrest. On the other hand, it will increase tumorigenesis 41. Sivonova et al reported that p53 and p21 polymorphisms are associated with pc 42. Eastham JA et al reported up-regulation of CDKN1A inhibited PC growth both in vitro and in vivo 14. Therefore, we hypothesize that the role of CDKN1A tumor suppressor is probably a functional mechanism of TH in treating PC.
FOS is another key up-regulated gene in response to TH in the PC cell line that acts as a tumor suppressor gene during the progression and invasion of PC and in the prominent mesenchymal transmission pathways K Ras, JAK-STAT, and epithelial 43. Shankar E et al reported that androgenic control of c-FOS can be introduced as a targeted therapy in PC 44. We suggest that TH can probably inhibit the progression and invasion of PC by up-regulating the FOS gene.
CDC20 is a hub-bottleneck up-regulated protein in PPIN. Abnormal expression of the Cell Division Cycle20 (CDC20) is associated with malignant progression and poor prognosis of cancers. CDC20 activates the anaphase-stimulating cyclosome complex (APC/C) which increases mitotic output through protein proteasome degradation and ubiquitination, and inhibits metastasis, increasing cell cycle arrest 45,46. Liang Dai et al reported that CDC20 contributes more to progression and migration in metastatic Prostate Cancer (mPCa) compared with PC and can be used as a therapeutic target in mPCa 47.
MDM2 can ubiquitinate the P53 protein and deliver it to the proteasome. Its overexpression is associated with resistance to chemotherapy in malignancies 48. Inhibition of MDM2 leads to P53 activation and can inhibit growth and metastasis independent of P53 and androgen receptors 49. Leite KR et al reported abnormal expression of oncoprotein MDM2 in PC. This study showed overexpression of MDM2 has a role in prostate carcinogenesis 50.
CCNB1 and CCNB2 are type B cyclins and are essential components of the cell cycle regulatory system. CCNB2 is linked to the Fox and P53 signaling pathways and is a critical enzyme that mediates the progression of cell cycle-related oxidation. In the FoxO signaling pathway, CCNB2 may bind to TGF β-R II and be involved in growth factor-b-conversion and mediated cell cycle control 51. Gomez LA et al in 2007 reported a positive correlation between cyclin B1 protein and inducing apoptosis 52. So, Shen H et al showed CCNB1 and CCNB2 genes are key factors in cell cycle pathways and the development of PC. These factors can suggest a novel prognosis and treatment bio-markers 53. The molecules (POLQ, CDC20, MDM2, CCNB1, and CCNB2) have shown a slight increase in the LNCaP cell line treated with TH but studies have shown an increase in the expression of these molecules is associated with the progression of PC. Therefore, an experimental investigation is necessary to validate their expression and explain their possible roles in the treatment of PC with TH.
It has been shown that TH has effects on functional mechanisms, including response to oxygen levels, regulation of cell cycle, response to immobilization stress, response to a toxic substance, regeneration, p53 signaling pathway, parathyroid hormone synthesis, secretion and action, and FoxO signaling pathway. Low oxygen levels characterize prostate tumors, hypoxia is associated with prostate tumor progression 54. In hypoxia, prostate tumor cells are overly resistant to chemotherapy drugs due to mutation or inactivation of P53 or BCL2. Drug resistance can be partially attributed to increased multidrug resistance carriers, including P-glycoprotein (PGP) 55. Jing X et al in 2019 reported that hypoxia targeting might be effective in cancer treatment 56. So in 1991, Teicher BA et al showed a relationship between tumor oxygenation and response to radiation which could improve cancer treatment 57. Therefore, we hypothesize that TH can decrease the resistant drugs and increase apoptosis by the biological process of response to oxygen levels.
The other functional mechanism of this drug is regulating the cell cycle. The mechanism for anti-mitotic medicines is to disrupt the mitotic spindle complex, which activates the spindle assembly checkpoint, causing mitotic cessation and subsequent death of tumor cells in mitosis or slipping into the post-mitotic phase and exiting from mitosis. Levrier C et al in 2018 reported that TH indirectly inhibits tubulin-associated protein and regulates the cell cycle 4. Our study showed that TH in addition to proliferation can regulate repair, apoptosis, and metastasis in PC. However, experimental studies are needed to validate our results. We should also consider that in addition to cancer cells, normal cells have a high proliferative capacity and this drug may affect normal cells as well.
One of the reasons for resistance to cancer treatment is the continuous activation of the nervous system caused by stress, accelerating tumor growth through β2 adrenergic receptor signaling. Watabe T et al showed that the activation of β2 adrenergic receptor signaling by increased epinephrine in response to immobilization stress in PC delays the expression of drug-induced apoptosis-regulating proteins (MCL1) 58. On the other hand, androgen-induced maturation and regeneration are associated with the growth and metastasis of cells in PC 59,60. Therefore, we predict that drug could affect PC treatment by regulating these mechanisms.
P53 is also a tumor suppressor gene whose mutation is associated with more significant resistance to treatment. Rahman MA et al showed that although P53 typically inhibits autophagy, it has a dual role as a positive or negative regulator of autophagy in cancer. It may travel to the nucleus in response to stress or hunger and activate autophagy 61. Autophagy deregulation is a feature of tumor progression and its suppression with chemical treatments of potential cancer particles 62. Therefore, it can be concluded that TH activates autophagy in PC through P53 mediation. The experimental investigation is necessary for the validation of this mechanism.
The synthesis, secretion, and action of the parathyroid hormone are functions regulated by this drug. Cancer progression and mortality are also associated with elevated serum levels of Parathyroid Hormone (PTH), vitamin D, and calcium metabolism 63. PTH-related protein (PtHrP), a bone metastasis agent, is reported to produce the phospholipase activation pathway (PLC/PLA2/PLD) from IP3, which in turn activates IP3R (inositol 1, 4, 5-trisphosphate receptors). The other role of this hormone is to regulate calcium levels in the blood. Studies on its function related to PTEN in the transport of calcium from the cytosol to the mitochondria have shown that the competitive binding of these components regulates calcium secretion and controls apoptosis 64. Schwartz GG et al showed that increased expression of the PTH receptor and PTH promotes PC cells' growth and invasion to the bone 62. Brun VH et al, in 2021, reported that severe hypercalcemia generated by parathyroid hormone has a critical role in rectal cancer metastasis 65. Therefore, one of the TH roles can be regulating parathyroid hormone levels and preventing metastasis. The reduction of parathyroid hormone is associated with hypocalcemia which causes muscle cramping, abdominal pains, and paresthesia 66. These indications can be considered as side effects of the drug. Given the importance of this mechanism and the lack of experimental studies about its mediation in response to TH, we offer experimental investigation to identify the role of this factor in PC treatment.
Fork Head Box (Foxs) has many effects on stem cell growth, metabolism, and maintenance. Also, it acts as a tumor suppressor in the regulation of essential genes for cell proliferation, cell death, angiogenesis, and cell migration and metastasis 67. The loss of FOXO proteins function is critical in tumorigenesis 68. Therefore, this drug can activate the FoxO signaling pathway and decrease growth and metastasis in PC. Jiramongkol et al showed that genetic disorders and metabolic diseases could cause the deregulation of the FoxO signaling pathway. These disorders can be introduced as side effects 69. In this study, we evaluated the relationship between critical genes and PC using GEPIA2 datasets. The boxplot represented significant over-expression of CDC20 and CCNB1 in PC compared to normal prostate tissue (Figure 5). Besides, the overall survival indicated that among critical genes a decrease in CDKN1A and MDM2 expression and overexpression of EGR1, FOS, and CCNB1 is associated with reduced survival ability in patients.
We suggest that TH regulates PC proliferation, repair, apoptosis, and metastasis. The muscle cramping, abdominal pains, and paresthesia regulated by the parathyroid hormone, and metabolic diseases regulated by FoxO signaling pathway have probably side effects mediated by this drug in our model. The expression of some genes that have an important role in the proliferation and metastasis of PC, increases in the cells treated with this drug, therefore, in vitro and in vivo studies are required to validate our results.
Conclusion :
This study used a network-based approach (PPIN and GRN) to identify hubs-bottlenecks, 3-nodes motifs consisting of target genes, miRNAs, and TFs, and molecular mechanisms and pathways in PC in response to TH. POLQ, EGR1, CDKN1A, FOS, MDM2, CDC20, CCNB1, and CCNB2 were determined as critical DEGs, mediating response to TH in PC. Response to oxygen levels, regeneration, immobilization stress, and a toxic substance, regulation of cell cycle, p53 signaling pathway, synthesis, secretion, and action of parathyroid hormone, and FoxO signaling pathway are essential biological processes and functional pathways related to TH in the PC. Besides, muscle cramping, abdominal pains, paresthesia, and metabolic diseases were determined as pathways possibly mediating its side effects. We recommended in vitro and in vivo investigation for validation of our results.
Acknowledgement :
This study is related to project NO.1399/62246 of the Student Research Committee, Shahid Beheshti University of Medical Sciences, Tehran, Iran. We also appreciate the "Student Research Committee" and "Research and Technology Chancellor" at Shahid Beheshti University of Medical Sciences for their financial support of this study.
Conflict of Interest :
The authors declare no conflicts of interest.
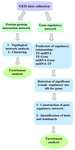
Figure 1. Workflow study.
|
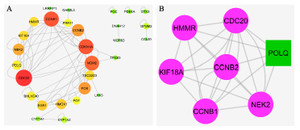
Figure 2. PPI network and MCOD cluster. A) The nodes with a high degree are shown with a bigger size and the color of dark red. B) The green node represents the seed gene.
|
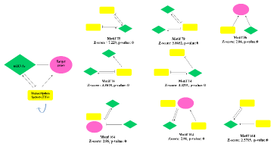
Figure 3. The regulatory motifs are consisting of genes, TFs, and miRNAs were detected by FAN MOD software with their Z-score and p-value. Four relationships miRNA-gene (miRNA represses gene expression); miRNA-TF (miRNA represses TF gene expression); TF-gene (TF regulates genes expression); and TF-miRNA (TF regulates miRNA expression).
|

Figure 4. Gene regulatory network. The shared genes of 10% highest degree, betweenness centrality, and closeness centrality merged motifs. The yellow rectangle is transcription factors, the green diamond show miRNAs, and the pink circular represents the genes.
|

Figure 5. The boxplot revealed the different expression levels of critical genes in prostate cancer rather than normal samples (GEPIA2). PARD, prostate adenocarcinoma.
|
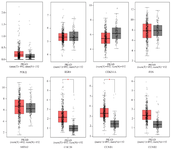
Figure 6. The Kaplan-Meier plot shows the eight critical genes effects on the overall survival rate of prostate cancer patients (by GEPIA2). Low and high TPM are represented in all graphs.
|
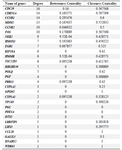
Table 1. The list of genes with degree >1 obtained with Network Analyzer plugin in Cytoscape software.
|
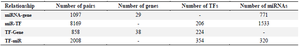
Table 2. Summary of four types of regulatory relationships among miRNA-gene, TF-Gene, miR-TF, and TF-miR interactions
|
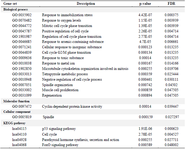
Table 3. The biological processes, molecular function, cellular components, and KEGG pathways identified using the Webgestalt tool (p<0.05)
|
|