A New Specific DNA Target Sequence for Identification of Staphylococcus epidermidis using Modified Comparative Genomic Analysis
-
Khoshbakht, Reza
-
Antimicrobial Resistance Research Center, Mashhad University of Medical Sciences, Mashahd, Iran
-
Student Research Committee, Mashhad University of Medical Sciences, Mashahd, Iran
-
Zare, Hosna
-
Antimicrobial Resistance Research Center, Mashhad University of Medical Sciences, Mashhad, Iran
-
Student Research Committee, Mashhad University of Medical Sciences, Mashhad, Iran
-
Kamali Kakhki, Reza
-
Antimicrobial Resistance Research Center, Mashhad University of Medical Sciences, Mashhad, Iran
-
Student Research Committee, Mashhad University of Medical Sciences, Mashhad, Iran
-
Neshani, Alireza
-
Student Research Committee, Mashhad University of Medical Sciences, Mashhad, Iran
-
Department of Laboratory Sciences, School of Paramedical Sciences, Mashhad University of Medical Sciences, Mashhad, Iran
-
 Arfaatabar, Maryam
Department of Medical Laboratory Sciences, Kashan Branch, Islamic Azad University, Kashan, Iran, arfahmara081@gmail.com
Arfaatabar, Maryam
Department of Medical Laboratory Sciences, Kashan Branch, Islamic Azad University, Kashan, Iran, arfahmara081@gmail.com
-
Department of Medical Laboratory Sciences, Kashan Branch, Islamic Azad University, Kashan, Iran
Abstract: Background: Staphylococcus epidermidis (S. epidermidis) is the most frequently isolated pathogen from prostheses infections in the body. Therefore, improving its diagnostic methods, including rapid Nucleic Acid Amplification Tests (NAAT), seems necessary. Since the first step in designing a NAAT is to find a specific sequence and all DNA targets that have been introduced so far are not completely specific, we introduced a new 100% specific DNA target sequence to identify S. epidermidis in this study.
Methods: Modified comparative genomic analysis was used to find the best specific target sequence to detect S. epidermidis. A PCR method was designed for the evaluation of this target. To determine the detection limit and analytical specificity, pure genomic DNA of 18 bacteria include 12 standard strains (one S. epidermidis and 11 non-S. epidermidis) and six clinical isolates (five S. epidermidis and one non-S. epidermidis) were used.
Results: The 400 bp sequence of S. epidermidis ATCC 14990 was identified as the most specific sequence (Se400), having a 100% sequence similarity to S. epidermidis genomes but not with other bacteria. The detection limit of Se400-PCR was 10 fg, equal to about 4 copies of S. epidermidis genomic DNA/μl. All pure DNA templates from S. epidermidis generated a detectable amplicon by 264 bp length, but the PCR test was negative for the non-S. epidermidis group.
Conclusion: The Se400 sequence can be considered as a specific target for detecting S. epidermidis, based on our findings.
Introduction :
One of the most isolated members of the coagulase-negative staphylococci (CoNS) group is Staphylococcus epidermidis (S. epidermidis). This bacterium colonizes mucous membranes and the skin, accounting for the majority of the bacterial flora in this environment 1-3. Genome study of S. epidermidis revealed that it is fully equipped with genes supposed to offer resistance from the severe circumstances faced in surrounding environment, allowing it to remain longer in dry conditions in hospitals 4. S. epidermidis is the most commonly implicated pathogen in infections related to any form of an indwelling medical device 5. This micro-orga- nism has been detected with a relatively high preva-lence from the Central Nervous System (CNS) shunts, joint prostheses, and prosthetic valves 6,7. Also, the mentioned bacterium has been repeatedly isolated from different specimens, such as blood, skin, wound, urinary tract, soft tissue infections, endocarditis, bacteremia, and pneumonia 8,9. According to scientific documents, CoNS cause half of all cases of Prosthetic Valve Endocarditis (PVE) 10. More than 20% of people with implanted cardiac devices are infected by S. epidermidis, which in turn causes pain and purulence at the infection site and sepsis 6. The sepsis mortality rate resulting from S. epidermidis in infants could be as high as 4.8 and 9.4% 11. The mortality rate due to endocarditis caused by CoNS is reported to be about 36% 6, while it is estimated at 30% for septic shock 12.
Traditionally, S. epidermidis diagnosis has been performed according to the biochemical tests and morphological characteristics. Such methods are time-consum-ing (sometimes up to several days) and do not reliably distinguish S. epidermidis from other CoNS. Therefore, finding faster and more reliable methods has always been required. The development of Nucleic Acid Amplification Tests (NAATs) such as Polymerase Chain Reaction (PCR) in recent decades, has greatly increased the speed, sensitivity, and specificity of diagnostic tests 13-15. One of the critical points for designing a NAAT is a completely specific DNA sequence for the desired micro-organism. The specific sequence should be present in all strains of such micro-organism but not found in any other micro-organism or has very low similarity 16. In recent years, various PCR tests have been designed to detect S. epidermidis based on genes such as serp0107, gseA, Staphostatin A, and sesC 17-20. Our bioinformatics evaluation showed that all genes introduced so far as diagnostic targets, are not 100% specific, and have many similarities with other species of Staphylococcus. Therefore, finding a specific target sequence that can be applied to design a completely specific PCR is still needed. Modified comparative genomic analysis or modified genome comparison is among the methods for finding a specific target sequence for organisms and was introduced by our team in 2018 16,21. So, this study aimed to introduce a novel target gene that is specific for the S. epidermidis complex, as well as to design highly specific and sensitive primers for the rapid detection of S. epidermidis using modified comparative genomic analysis.
Materials and Methods :
Staphylococcus epidermidis-specific target mining
According to the described method in our previous studies 16,21, the genomic sequence of S. epidermidis ATCC 14990 was compared with the available genomic sequences on the nucleotide collection database 22,23, and the most specific sequence was selected (Figure 1). The steps are described below:
1. Genomic sequences of S. epidermidis on nucleotide collection database were determined. Then, one case which was preferably the NCBI reference sequence, was regarded as the reference.
2. The sequence of S. epidermidis ATCC 14990 (NZ_CP035288.1) was selected as the reference, and the sequence was obtained and cut to 5000 bp independent fragments, producing about 493 fragments.
3. Separately, each fragment was compared with other available sequences of nucleotide collection database by Basic Local Alignment Search Tool (BLAST). BLAST discovers similar regions between DNA sequences. The nucleotide sequences is compared with available sequences on database and the statistical significance is calculated by the program 22.
4. After each analysis, results were screened, and the best fragments were selected. Evaluation of results was performed by two criteria:
a. Presence of all S. epidermidis NCBI reference sequences in search results, having both identity and query cover of 100%.
b. No other microorganism except S. epidermidis would appear with the query cover >90%. The selection of these two criteria was based on our experiences and evaluation of the first 200 fragments.
5. Selected fragments of the previous step were compared separately with non- S. epidermidis sequences of the nucleotide collection database, and conserved parts of each fragment were determined.
6. Finally, the longer specific part was selected, and we named it Se400.
Primer design and PCR
To evaluate the specificity of Se400, an end-point PCR was designed with Oligo7 software 24. The primers were then tested for secondary structure and anticipated melting temperature using Oligo Analyzer 3.1 (https://eu.idtdna.com/calc/analyzer) and were manufactured by DENAzist Asia Company. 264 bp amplicon and Primer sequences are provided in table 1. PCR reaction was prepared in 25 μl containing 2.5 μl of 10×PCR buffer (100 mM Tris-HCl [pH=8.3], 500 mM KCl), 1 μl of each 10 μM forward and reverse primers, 1 μl of DNA sample, 0.5 μl of 200 μM (each) of the four dNTPs, 1.5 μl of 25 mM MgCl2, 0.625 U of Taq DNA polymerase, and PCR grade water. 10 ng of S. epidermidis ATCC 14990 pure DNA was used as the positive control, and water as the negative control.
For DNA amplification, 5 min of initial denaturation at 95°C was followed by 30 cycles of (i) 45 s of denaturation at 95°C, (ii) 45 s of annealing at 49°C for, (iii) 60 s of extension at 72°C for, and (iv) final 5 min of extension at 72°C. Finally, 3 μl of PCR product was visualized using 1.5% agarose gel electrophoresis and DNA green viewer. The presence of a 264 bp amplicon specifies the positive result.
Bacterial isolates
In this study, pure genomic DNA of 18 bacteria, including 12 standard strains (one S. epidermidis and 11 non-S. epidermidis) and six clinical isolates (5 S. epidermidis and one non-S. epidermidis) were used. All standard strains were acquired from the microbial bank in the Antimicrobial Resistance Research Center of Mashhad University of Medical Sciences, and all clinical isolates were acquired from Imam Reza Hospital of Mashhad (Table 2).
Analytical sensitivity (limit of detection)
The pure DNA concentration of S. epidermidis ATCC 14990 was calculated by a spectrophotometer (Thermo Scientific). A serial dilution of pure DNA was then prepared in water (10 ng, 1 ng, 100 pg, 10 pg, 1 pg, 100 fg, 10 fg, 1 fg) and the volume of 1 μl was applied as the template. The process was repeated three times to ensure the results.
Analytical specificity
The primer specificity was investigated by Blastn software to determine cross-reactivity with other human or bacterial genomes. To determine the analytical specificity for Se400-PCR, pure DNA of six S. epidermidis and 12 non-S. epidermidis were used (Table 2). 10 ng of pure DNA was applied in each reaction. Finally, sequencing was performed on all positive PCR products.
Results :
Target mining
The 400 bp sequence containing nucleotides 242, 200 to 242,600 of S. epidermidis ATCC 14990 (NZ_ CP035288.1) was recognized as the most specific sequence, having a 100% sequence similarity to S. epidermidis genomes but not with other bacteria. Se400 is a non-coding sequence located between EQW00_ 01195 and EQW00_01200 genes (Figure 2). The blastn search showed that Se400 could detect all strains of S. epidermidis among the registered complete genomes in the nucleotide collection database, and no similarity was observed with other micro-organisms.
Analytical sensitivity (limit of detection)
The detection limit is defined as the lowest analyte concentration that can be reliably detected. Consequently, the Se400-PCR detection limit was 10 fg, equal to about 4 copies of S. epidermidis genomic DNA/μl (Figure 3).
The analytical specificity
PCR amplification using Se400-specific primers was performed with 18 bacteria using pure genomic DNA as the template to consider the analytical specificity of the Se400-PCR. As presented in figure 4, all genomic DNA templates from S. epidermidis generated a detectable amplicon by 264 bp length, but the PCR test was negative for the non-S. epidermidis group; subsequently, the Se400 primer was specific for detecting S. epidermidis. Finally, analysis of sequencing results for PCR products from 6 positive samples showed that the produced amplicon is related to the Se400 sequence of S. epidermidis.
Discussion :
Before the discovery of molecular methods, phenotypic and biochemical tests were the only powerful methods for the detection and differentiation of various bacteria, including S. epidermidis. With the development of molecular methods, especially PCR in recent decades, the problems of traditional methods such as being slow and lacking reliability were solved 13,14. One of the challenges for designing a PCR test to detect S. epidermidis is the lack of a completely specific DNA target. According to the literature, most NAATs in the 1990s used genus-specific targets due to the unavailability of species-specific DNA targets. The amplified sequences based on such genes could only differentiate S. epidermidis from other close species when they were analyzed by confirmatory methods. The most famous method for analyzing these fragments was sequencing and then comparing the results with available sequences on GenBank. 16S rDNA, sodA, hsp60, and tuf were among the most used genes in such methods 25-28. Although the addition of the sequencing step reduced the speed and increased the costs, some cases were still seen in which the sequenced fragment was quite similar in several species 25,29. For example, although the 16S rDNA gene has been suggested as a target gene, it cannot be used as a distinct target to detect S. epidermidis in clinical specimens due to the significant similarity to Staphylococcus aureus (S. aureus) 30-32.
With an increased detection of S. epidermidis infections in the last two decades, the need to find species-specific targets for the detection of this bacterium has been increased significantly. Efforts eventually led to the introduction of serp0107, gseA, ecpB, and SesC genes as species-specific targets 17-20. However, our bioinformatics evaluation before starting this project showed that none of these diagnostic targets was 100% specific (Table 3).
In 2004, the gseA gene (GenBank acc. No. AB096695), responsible for the production of glutamic acid-specific 27-kDa serine protease (GluSE), was introduced by Ikeda et al as a species-specific target of S. epidermidis. This protease is involved in degrading human fibronectin, collagen, the complement protein C5, and slime formation. Thus, the protease may be associated with the pathogenesis of S. epidermidis 17,33. Despite an appropriate length of this gene (1214 bp), a comparison of its sequence with other available sequences at nucleotide collection database showed that this gene has similarity with some parts of the Staphylococcus saccharolyticus (S. saccharolyticus) genome with >68% query cover and >75% identity. This bacterium is a normal flora of the skin, and the pathogenesis is not apparent yet. Furthermore, very similar sequences to the gseA gene are present in other bacteria, reducing the specificity of this gene for the detection S. epidermidis. These bacteria include Staphylococcus capitis (S. capitis), Staphylococcus caprae (S. caprae), S. aureus, Staphylococcus haemolyticus (S. haemolyticus), Staphylococcus saprophyticus (S. saprophyticus), Staphylococcus hominis (S. hominis), Staphylococcus equorum (S. equorum), Staphylococcus simulans (S. simulans), Staphylococcus lugdunensis (S. lugdunensis), and Staphylococcus schleiferi (S. schleiferi) serp0107, which is a putative transcriptional regulator gene, was introduced by Liu et al in 2006 as the species-specific diagnostic target to detect S. epidermidis. The sequence of this gene with 882 bp length was obtained from the (nt 91276–92157) of S. epidermidis RP62a (GenBank Accession No. CP000029) 18. Our assessment of this sequence using the BLASTN search tool showed that a highly similar sequence is also present in S. saccharolyticus and S. capitis. Also, similar points to the serp0107 gene are found in other Staphylococcus species, including S. caprae, S. haemolyticus, Staphylococcus warneri (S. warneri), S. hominis, and Staphylococcus argenteus (S. argenteus) leads to a lack of complete specificity of this gene for the detection of S. epidermidis.
S. epidermidis surface protein C (SesC) gene was introduced by Khodaparast et al in 2016 as a specific diagnostic target of S. epidermidis. SesC protein is expressed more in S. epidermidis biofilm-associated cells than planktonic ones. Also, this target is appropriate to design various NAATs due to the suitable length (2031 bp) 19. Nevertheless, our evaluation of this gene showed that similar sequences are found in three other species of the Staphylococcus genus, which reduces its specificity. The highest similarity was seen for the S. saccharolyticus with 83-100% query cover and >69% identity, followed by S. capitis and S. caprae.
Finally, the last introduced species-specific gene for this bacterium was the ecpB gene, encoding the Staphostatin A protein. This 318 bp gene was initially introduced to differentiate S. aureus and S. epidermidis 20. However, the high similarity with a sequence in S. aureus with 65% query cover and 67% identity makes it unspecific. The assessment of available sequences in NCBI also showed that this gene is also found in S. saccharolyticus with 100% query cover and >77% identity.
According to the results obtained using the BLA-STN search tool, the Se400 sequence is completely specific to the S. epidermidis and is not found in any other micro-organisms, unlike the other sequences having nonspecific regions in several points. Therefore, using this sequence seems to solve the unavailability of a completely specific target for S. epidermidis.
In this study, the specificity of the Se400 target sequence was confirmed by PCR. The specificity of Se400 in all S. epidermidis strains highlighted both the efficacy of the comparative genomic analysis for finding possible targets and the significance of experimental research. Also, the Se400-PCR test was able to detect very low levels of genomic DNA template. One of our limitations was the small number of bacterial strains tested, and it is recommended that further research be conducted with a large sample size. Future studies might include comparing the previously known target gene with Se400 sequence in clinical samples.
Conclusion :
In conclusion, S. epidermidis-specific target sequences were identified using a new comparative genomics method for finding species-specific nucleotide sequences. Many nucleotide targets were assessed, and a target sequence was applied to design a PCR test to detect S. epidermidis in clinical samples. Further surveys are being planned to include more bacterial strains for the evaluation of the particular targets. Unique targets may be found using this method for the detection of any micro-organism, for which a genome sequence is available.
Conflict of Interest :
None declared.
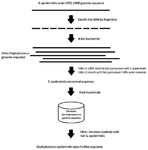
Figure 1. Method for mining S. epidermidis-specific nucleotide sequences.
|

Figure 2. Schematic location of Se400 between EQW00_01195 and EQW00_01200 genes.
|
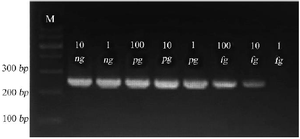
Figure 3. Gel electrophoresis of Se400-PCR product on 1.5% agarose gel. 264 bp amplicons are shown at various concentrations of S. epidermidis pure DNA as template. Amplicons using 10 ng, 1 ng, 100 pg, 10 pg, 1 pg, 100 fg, 10 fg, and 1 fg are shown in lanes.
|

Figure 4. Gel electrophoresis of Se400-PCR products. The amplicon size is 264 bp. M: 100-bp molecular DNA ladder (Pars Tous, Iran).
|
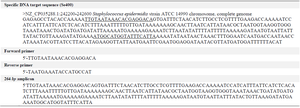
Table 1. The Se400 sequence, the designed primers for PCR, and amplicon
|

Table 2. The list of bacteria used in this research
|
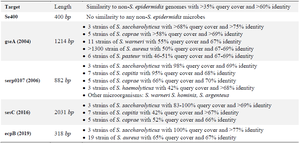
Table 3. Comparison of Se400 sequence specificity with other introduced genes for detecting S. epidermidis in species level
The specificity of the genes was tested bioinformatically using Blastn software.
|
|