Expression of Toll-Like Receptors 2, 4 and 5 in Relation to Gut Microbiota in Colon Neoplasm Patients with and without Inflammatory Bowel Disease
-
Asadzadeh Aghdaei, Hamid
-
Basic and Molecular Epidemiology of Gastrointestinal Disorders Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
-
 Rezasoltani, Sama
Foodborne and Waterborne Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran, samasoltani70@gmail.com
Rezasoltani, Sama
Foodborne and Waterborne Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran, samasoltani70@gmail.com
-
Foodborne and Waterborne Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
-
Olfatifar, Meisam
-
Gastroenterology and Liver Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
-
Nazemalhosseini Mojarad, Ehsan
-
Gastroenterology and Liver Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
-
Sherkat, Ghazal
-
Basic and Molecular Epidemiology of Gastrointestinal Disorders Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
-
Yadegar, Abbas
-
Foodborne and Waterborne Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
-
Feizabadi, Mohammad Mehdi
-
Department of Microbiology, School of Medicine, Tehran University of Medical Sciences, Tehran, Iran
-
Zali, Mohammad Reza
-
Gastroenterology and Liver Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Abstract: Background: Toll-Like Receptors (TLRs) are the critical mediators of inflammatory routs in the gut, which play an essential role in regulating the immune responses towards various ligands derived from pathogenic bacteria. Also, TLR signaling has been implicated in the development of Inflammatory Bowel Disease (IBD), Adenomatous Polyp (AP), and Colorectal Cancer (CRC). Here, we aimed to examine the expression of some TLRs concerning certain fecal bacteria in AP and CRC patients with and without IBD.
Methods: This case-control study collected fecal and colonic tissue samples from 93 patients versus Normal Controls (NC) via colonoscopy. Fecal samples were used for DNA extraction, and the abundance of selected fecal bacteria was determined by absolute real-time PCR. Also, the gene expression of TLR2, 4, and 5 was analyzed using RT-PCR on the colonic tissues of participants.
Results: Compared to NC individuals, in AP and CRC patients, the mRNA expressions of TLR4 and TLR2 were significantly increased while TLR5 was decreased. A meaningful association between TLRs mRNA expression levels and the abundance of some selected fecal bacteria was detected. Also, there was a significant relationship between participant’s food regimes, smoking habit and intestinal TLRs expression.
Conclusion: Our study proposed the important role of TLRs during adenomatous and CRC formation. Alterations in TLRs expression associated with certain gut bacteria may contribute to disease development.
Introduction :
Colorectal Cancer (CRC) has been regarded as one of the most prevalent cancers worldwide, and its incidence is predicted to increase in the following years. Despite significant improvement in CRC treatment, its mortality rate remains high, and a great number of CRC patients are at an increased risk to die 1. CRC arises because of environmental and genetic factors cooperating to form colon polyps that may progress to malignancy 2. Some risk factors such as age, race, he-redity, obesity, smoking, and alcohol consumption canalter colorectal epithelial change that can lead to tumor development 3. In terms of genetic factors, three path-ways comprise mutations in DNA mismatch repair genes indicated as Microsatellite Instability (MSI), mutations in Adenomatous Polyposis Coli (APC), and other genes that activate the Wnt pathway represented by Chromosomal Instability (CIN) phenotype, and global genome hypermethylation marked as CpG Island Methylator Phenotype (CIMP), can be characterized in CRC patients 4. Epigenetic regulation of gene expression is a general pivotal mechanism that is effective in normal tissues and essential in preserving genomic stability, embryonic development, and tissue differentiation 5. Moreover, chronic or repeated episodic inflammatory insults to the intestinal mucosa have a clear mechanistic role in the development of Inflammatory Bowel Disease (IBD)-associated CRC 6,7. The polyp to cancer development sequence is primarily driven at the cellular level by gene mutations and epigenetic alterations, which is identified to be a heterogeneous process 1,2. Environmental factors such as gut microbiota have significant role in these epigenetic alterations 8,9. Gut microbiota have an important function in the biological microenvironment; more than 15% of cancers have been known to be developed by microbiota and those linked with liver and gastrointestinal tract are certainly recognized as being associated with microbes 9,10. Moreover, the epigenetic modification of oncogenes, tumor suppressors, miss match repair genes, and proinflammatory mediators are shown to be involved in disruption of homeostatic balance and induction of dysbiosis 8.
Also, gut microbiota is associated with mediating CRC progression by their structural antigens that interact with host’s Microbe-Associated Molecular Patterns (MAMPs) and Pathogen-Associated Molecular Patterns (PAMPs) receptors such as Toll-Like Receptors (TLRs) 11. TLRs detect pathogenic bacteria, identify intracellular anomalies, and mount an immune answer, hence play a major role in immune system homeostasis 11,12. Unusual activation of TLRs imperils ordinary physiological processes and results in different types of inflammatory diseases such as IBD, cancers, and autoimmune disorders 13,14. In similar fashion, TLRs are identified to play an important role in CRC that affects the colon 15. Large intestine and rectum are completely inhabited by microbiota, emphasizing the serious role of TLRs in disease pathogenesis 16.
To maintain colon homeostasis both locally and systemically, tolerance should be achieved by the induction of anti-inflammatory molecules 15,16. Commensal microbiota are vital regulators of the immune system and can maintain homeostasis by stimulating antibody production and activating immune cells 17-19. Herein, we tried to determine the relative expression levels of TLR2, TLR5, and TLR4 (some critical TLRs during CRC initiation and progression) in intestinal biopsy specimens of patients with Adenomatous Polyp (AP) and CRC with or without IBD compared to normal participants. Also, certain gut bacteria in the matched stool samples from these individuals were quantitatively evaluated to investigate the association between TLRs and gut microbial patterns for the first time. We found that intestinal expression of TLR2, TLR4, and TLR5 is not constant and depends on the commensal gut micro bacterial pattern. Also, we observed a significant association between participants’ food regimes, smoking habits, and intestinal TLRs expression.
Materials and Methods :
Data collection for study population
Participants of the study population included 42 patients with AP, 20 CRC cases with and without IBD and 31 Normal Controls (NC) and were enrolled between 2018 and 2019 in Taleghani Hospital, where they had been referred for CRC screening. The study population only included individuals without previous colon or rectal surgery, CRC and infectious injuries of the large intestine and rectum 20,21.
Tissue sample collection
Colonic tissue samples were collected from the participants during the colonoscopy procedure. Written consent and the study protocol were approved by the Clinical Research Ethics Committee of the Tehran, Iran.
Fecal sample collection
Fresh stool samples were collected two weeks to three days before colonoscopy from all individuals prior to bowel clean. Whole fresh stool was collected in sterile boxes, and 10 g was frozen at -20°C. All samples were then selected for DNA extraction and absolute quantitative RT-PCR.
RNA extraction from tissue samples
RNA was extracted from the tissue samples by using TRIzol reagent and stored at -70°C for further analysis.
DNA extraction from stool
Genomic DNA was extracted from frozen fecal samples using QIAamp DNA Stool Mini Kit (Qiagene, Hilden, Germany) per the manufacturer’s instructions.
TLR2, TLR4 and TLR 5 expression analysis in tissue samples
RNAs were converted to cDNA by reverse transcription reaction using Primescript TM RT Reagent kit (Takara, Japan). Following cDNA synthesis, real-time PCR and relative quantification method were performed using Premix Ex Taq SYBR (Takara, Japan) to evaluate the TLR2, TLR4 and TLR5 gene expression level. Real-time PCR amplifications were carried out in a Rotor Gene system (QIAGEN, Germany) using previously described primers 22,23 the Following conditions were used for PCR amplifications: 95°C for 5 s, 40 cycles of 95°C for 5 s, 60°C for 34 s, 95°C for 15 s, 60°C for 1 s and 60°C for 15 s 2. Amplification signals were normalized to β-glubin reference gene. Rest software was applied to determine the gene expression level of TLRs using Pfaffl method 24.
Bacterial quantification in fecal samples
Fecal bacterial candidates were quantified in stool samples of participants by absolute qRT-PCR using SYBR green detection system. The selected target fecal bacteria included Streptococcus bovis/gallolyticus, Enterococcus faecalis (E. faecalis), Bacteroides fragilis (B. fragilis), enterotoxigenic Bacteroides fragilis (ETBF), Fusobacterium nucleatum (F. nucleatum), Porphyromonas gingivalis (P gingivalis), Porphyro-monas spp. and Roseburia spp. DNA amplification was performed in a final volume of 20 μl, SYBR Premix EX Taq II (2X) (TLi RNaseH Plus), 20 pmol of each forward and reverse primer, 2 μl of extracted DNA. The amplification program was 35 cycles of 95°C for 5 s and 60°C for 34 s with an initial cycle of 95°C for 10 min and a primer pair-specific annealing temperature for 60 s 2.
Statistical analysis
Descriptive statistics was used to analyze the gene expression level of TLR2, TLR4 and TLR5 in terms of demographic characteristic variables. The Pearson correlation test was applied to measure the association between the gut bacterial quantity and TLRs expression. Also, t-test and one-way ANOVA or their nonparametric equivalents were applied to compare the gene expressions in terms of baseline variables. Subsequently the post hoc test was applied for determining the probable significant relationships.
The determination of gut bacteria's quantity was done based on 16SrDNA duplication. Some of the primers for bacterial species were selected from previous studies using nucleotide-nucleotide BLAST software to control their specificity, while others were specifically for this study. The primes for TLR genes were selected from previous authentic studies and analyzed with nucleotide-nucleotide BLAST (Table 1).
Results :
According to the results obtained for TLRs mRNA expression level, lower expression level of TLR5 (p=0.033) and higher expression level of TLR2 and TLR4 (p=0.000) were detected among CRC cases compared to normal participants. Also, AP cases and control groups were compared and a decreased level of TLR5 (p=0.041) mRNA expression and increased level of TLR2 and TLR4 (p=0.003) were obtained in AP cases in contrast to normal subjects (Figure 1).
Intestinal bacterial quantification in stool samples of participants showed that the abundance of F. nucleatum (p<0.001), Porphyromonas spp. (p<0.001), P. gingivalis (p<0.005), and E. faecalis (p<0.001) was increased in CRC cases compared to AP patients, whereas the abundance of S. bovis (p<0.001), ETBF (p< 0.001), B. fragilis (p<0.015) statistically increased in CRC and AP cases compared to controls (Table 2).
The relationship between demographic characteristics of the participants and mRNA expression level of the studied TLRs were also interpreted (Table 3). Interestingly, significant relationships were detected between food regimes, smoking habit of participants and TLRs mRNA expression level.
The relationship between targeted TLRs mRNA expression level and selected gut bacterial abundance was evaluated in different study groups by Pearson regression analysis (Table 4). Surprisingly, there was a significant relationship between TLR2, TLR4 mRNA ex-pression levels and the abundance of Roseburia spp., P. gingivalis (p<0.05), and between TLR5 mRNA expression levels and the abundance of ETBF in normal participants. Moreover, significant association between TLR2 mRNA expression level and the abundance of S. bovis, ETBF, and TLR4 mRNA expression level and Roseburia spp. abundance were detected in AP cases. Moreover, significant relationship between TLR2 mRNA expression and S. bovis abundance, TLR4 mRNA expression and E. faecalis abundance were detected in CRC cases.
Discussion :
The human intestinal is the host of various microorganisms and maintains over a thousand species. Several factors such as age, sex, genetics, diet, and lifestyle have affected each person's gut microbial pattern 25. The host intestine supplies a nutrient-rich environment where microbiota can prosper and assist the host homeostasis modulation 26. Aberrant disruptive modifications in the gut microbiome profoundly contributed to the improvement of colorectal cancer. Gut microbiota may play a crucial part in the CRC's advancement through their metabolite or structural component interacting with the host intestinal epithelial cell 27. Also, the association between inflammation and colorectal tumor pathogenesis is becoming increasingly critical. Several studies have verified that TLRs as immune molecules could mediate inflammatory response, which can play an essential role in this process 15-17. In the present study, the gene expression level of the most important TLR genes, including TLR2, TLR4 and TLR5 were analyzed in biopsy samples of patients with AP and CRC compared to control participants. The expression levels of TLR2 and TLR4 were significantly increased in both adenomatous and CRC cases, however a higher expression level was detected in CRC patients (p<0.0001). According to the relative expression levels of these TLRs in both cases of AP and CRC, it was assumed that upregulation of TLR2 and TLR4 may occur in the first step of CRC initiation. Hence, TLR2 and TLR4 could be offered as appropriate candidate for characterization of the adenoma and CRC in the first step of CRC initiation 10. The present results also confirmed that TLR2 and TLR4, which have been shown to upregulate in the process of polyp and CRC development, can initially induce inflammation and increase the immune system stimulation against Gram-negative bacterial structures, bacterial components, and metabolites in the direction of more facilitated CRC progression 8.
On the other hand, we observed that TLR5 mRNA expression level was decreased in adenomas and CRC participants compared to normal group. This means that TLR5 mRNA expression level may be essential for maintaining balance and gastrointestinal hemostasis, and any imbalance or its downregulation could lead to intestinal inflammation and disease 28. Our results confirmed the Yang et al study, in which they demonstrated that TLR3 and TLR5 appear to be constitutively expressed in healthy gut, while TLR2 and TLR4 are produced at low levels 8. This finding suggests that TLR3 and TLR5 receptors expression is mediated to avoid autoinflammatory immune activation in response to indigenous microbiota. Also, our results were in agreement with the findings obtained by Kelly et al indicating that TLR3 and TLR5 appear to be constitu-tively produced in healthy gut, whereas TLR2 and TLR4 are expressed at low level 29. For instance, some Lipopolysaccharides (LPS) molecules are strong inducers of TLR4 signaling, while others are antagonistic 29,30, or the ability of some bacterial flagellins to prompt the TLR5, while other structures are non-activating 29. Furthermore, Kutikhin et al reported similar results to our findings regarding increased expression of TLR2 and TLR4 and decreased TLR5 mRNA expression among AP and CRC cases 28.
Lee et al demonstrated that the tumor formation's inhibition by B. fragilis, a bacterium with a defensive function against the development of experimental colitis in animals, in colitis-associated CRC relied on the production of the polysaccharide A from B. fragilis. The TLR2 signaling was responsible for the protective function of B. fragilis 31. Concerning metastatic CRC, Li et al found that a metastasis-related secretory protein cathepsin K (CTSK) is a crucial mediator between intestinal microbiota imbalance and CRC metastasis. They also identified that the tumor secreted CTSK could bind to TLR4 to prompt the M2 polarization of Tumor-Associated Macrophages (TAMs) through an mTOR-dependent pathway. CTSK's overexpression in human CRC tissues is always going along with high M2 TAMs in the stroma and is associated with CRC metastasis and poor prognosis 32.
Another TLR gene expression impact in CRC was investigated by Rezasoltani et al, who evaluated the mRNA expression of TLR9 in people with different colorectal polyps compared to the normal group to examine its expression level during CRC initiation. They found that aberrant surface expression of TLR9 on tumor cells may stimulate the growth and invasion of colorectal polyps leading to colorectal cancer initiation 33. In this work, we observed significant relationships between participant’s food regimes, smoking habit and intestinal TLR2, TLR4 and TLR5 expression. This means that, the consumption of vegetables, fruits, meats, processed food and being smoker or not has a critical effect on the expression level of certain intestinal TLRs expression and maintenance of the homeostatic balance.
In addition, the relationship between certain gut microbiota and intestinal TLRs expression levels was investigated among the study groups. We detected significant association between TLRs and the relative abundance of gut bacteria such as P. gingivalis, Roseburia spp., S. bovis, ETBF and E. faecalis. Such findings were similar to the past research which had proved that the interaction between gut microbiota and TLRs can affect host homeostasis and immune responses, and the alterations of gut microbiota can result in inflammation 20,34,35. This inflammatory response can regulate the interactions between endogens microbiota and TLRs, and are also essential for immune signals, intestinal epithelial cell proliferation, and gut homeostasis 2. Taken together, specific upregulation in intestinal tissue TLRs of CRC patients indicates that TLRs may play an important role in the prognosis of inflammatory disorders that may lead to malignancy and cancer.
This study has encountered some limitations. The first one was the small sample size. The second was that we collected the samples from the patients who had no colon or rectal surgery, CRC, and infectious injuries of the large intestine and rectum. Finally, we had a limitation in designing new oligonucleotide sequences for primers used, and most of the primers were selected from previous studies.
Conclusion :
Taking together, the current study showed upregulation in TLR2, TLR4 and downregulation of TLR5 in both AP and CRC cases and suggested that changes in TLRs expression may occur during AP and CRC formation and progression. TLR2 and TLR4 were upregulated and TLR3 and TLR5 were downregulated in all patients with AP and CRC compared to NC. Hence, TLR3 and TLR5 could be necessary for retaining in gut and have a protective duty vs. malignant transformation of the colon mucosa. The upregulation and downregulation of some TLRs in CRC tissues propose that TLRs may play an important role in the prognosis of inflammatory disorders that may lead to malignancy. Furthermore, this study demonstrated that the intestinal expression of TLR2, TLR4 and TLR5 is dynamic and depends on the presence of pathogenic and commensal gut microbiota. Hence, altered immune activation in response to dysbiotic microbiota may promote intestinal inflammation in a subset of patients with AP and CRC. Further investigations on TLR expression patterns and gut microbial interactions in CRC may help developing strategies in CRC screening.
Acknowledgement :
We are immensely grateful to the Taleghani Hospital's physicians, personnel, and patients collaborating in data collection for our study.
Conflict of Interest :
The authors declare no conflict of interest.
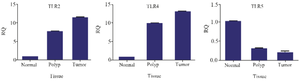
Figure 1. TLRs mRNA expression level in adenoma polyp and colorectal cancer patients with and without inflammatory bowel disease vs. normal control.
|
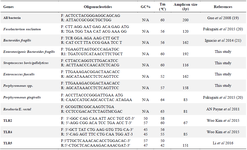
Table 1. Primers sequences in both gut bacteria and three TLR genes utilized in this study
|
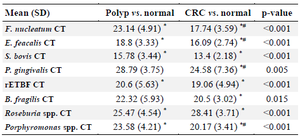
Table 2. Mean (SD) and p-value of each candidate bacterium based on CT interpretation in study groups
* p<0.05 vs. normal, # p<0.05 vs.
|

Table 3. The association between demographic characteristics of the participants and TLRs mRNA expression level
|
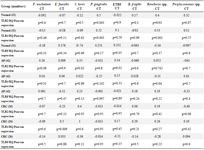
Table 4. The relationship between TLR2, TLR4 and TLR5 mRNA expression level and selected gut bacterial abundance in adenoma polyp and colorectal cancer patients with and without inflammatory bowel disease vs. normal controls
|
|