Exogenous Production of N-acetylmuramyl-L Alanine Amidase (LysM2) from Siphoviridae Phage Affecting Anti-Gram-Negative Bacteria: Evaluation of Its Structure and Function
-
Miri, Morteza
-
Department of Biotechnology, Faculty of Biotechnology, Semnan University, Semnan, Iran
-
Yazdianpour, Sepideh
-
Department of Biotechnology, Faculty of Biotechnology, Semnan University, Semnan, Iran
-
 Abolmaali, Shamsozoha
Department of Biology, Faculty of Basic Sciences, Semnan University, Semnan, Iran, Tel: +98 23 31532288 Fax: +98 23 31532220; E-mail: s_abolmaali@semnan.ac.ir
Abolmaali, Shamsozoha
Department of Biology, Faculty of Basic Sciences, Semnan University, Semnan, Iran, Tel: +98 23 31532288 Fax: +98 23 31532220; E-mail: s_abolmaali@semnan.ac.ir
Abstract: Background: To obtain endolysin with impact(s) on gram-negative bacteria as well as gram-positive bacteria, N-acetylmuramyl L-alanine-amidase (MurNAc-LAA) from a Bacillus subtilis-hosted Siphoviridae phage (SPP1 phage, Subtilis Phage Pavia 1) was exogenously expressed in Escherichia coli (E. coli).
Methods: The sequences of MurNAc-LAA genes encoding peptidoglycan hydrolases were obtained from the Virus-Host database. The sequence of MurNAc-LAA was optimized by GenScript software to generate MurNAc-LAA-MMI (LysM2) for optimal expression in E. coli. Furthermore, the structure and function of LysM2 was evaluated in silico. The optimized gene was synthesized, subcloned in the pET28a, and expressed in E. coli BL21(DE3). The antibacterial effects of the protein on the peptidoglycan substrates were studied.
Results: LysM2, on 816 bp gene encoding a 33 kDa protein was confirmed as specific SPP1 phage enzyme. The enzyme is composed of 271 amino acids, with a half-life of 10 hr in E. coli. In silico analyses showed 34.2% alpha-helix in the secondary structure, hydrophobic N-terminal, and lysine-rich C-terminal, and no antigenic properties in LysM2 protein. This optimized endolysin revealed impacts against Proteus (sp) by turbidity, and an antibacterial activity against Klebsiella pneumoniae, Salmonella typhimurium, and Proteus vulgaris in agar diffusion assays.
Conclusion: Taken together, our results confirmed that LysM2 is an inhibiting agent for gram-negative bacteria.
Introduction :
Cell wall protects the cells from the turgor pressure and helps maintenance of the cell shape in bacteria. Accordingly, every bacterial cell wall destructor is known as an antibacterial agent 1. Today, the cell wall hydrolyzing enzymes from bacteriophages targeting the peptidoglycan subunits have been the main concern because of their specific action and lack of drug-resistant effects. The peptidoglycan-hydrolyzing enzyme (Endolysin) as an antibacterial agent cleaves the linkage between peptide and carbohydrates in the peptidoglycan 2. Endolysins have several important advantages in comparison to antibiotics. The enzymes lyse bacterial cell wall within minutes to a few hours 3. They are active against both growing and dormant cells, disrupt bacterial biofilms, and kill drug-resistant strains 3. Bacteriophage-derived endolysins are potentially a long-term antibacterial replacement for antibiotics 3.
The catalytic and binding domains of endolysin are connected via a short linker 4. The enzymes attach to their target by the Cell wall Binding Domains (CBDs) and digest the peptidoglycan’s linkages through Enzymatically Active Domains (EADs) 5. Gram-negative and gram-positive bacteria differ in their endolysins’ structures. Gram-positive endolysins are composed of separate CBDs and EADs while single domain globular protein forms the most gram- negative endolysin 4,5. The endolysins cannot disrupt the cell membrane of gram-negative bacteria and only their structure and/or function can lead to outer membrane penetration 6. However, the enzymes either in combination with the membrane permeability or in conjugation with a small protein, holin, kill the gram-negatives. Holin makes a hole in the cell membrane of gram-negative bacteria 7.
Although the new sequencing methods and types of equipment have entailed a revolution in genome data, less than 1000 endolysins have been characterized 8. The limited knowledge about the mode of action plus the specific targeting of endolysins has sparked the idea to produce recombinant endolysins with desired features 8. They are suitable in two different aspects. The first, as mentioned above, is the specific digestion of the cell wall, which does not lead to antibacterial resistance impacts. The second is the possibility to engineer the endolysin domains for a new desired feature. Endolysin as a functional protein can be rearranged via normal genetic engineering methods whereas altering non-peptide antibacterial compounds is more difficult and complicated. Studying the evolution of phage lysins has revealed modular domain exchanges that lead to new catalytic binding properties 9. Novel endolysins can be developed with optimized stability, specificity, and lytic function 10.
Recently, recombinant endolysin as a bio-control agent has been investigated in a variety of gram-negative pathogens. Since N-acetylmuramyl L-alanine-amidase (MurNAc-LAA) specifically destroys the bacterial cell wall by cutting muramic acid-peptide bond 11, and muramic acid as a substrate is present in bacterial cell wall but not in higher organisms, amidase MurNAc-LAA was proposed as a biocontrol agent to limit the pathogenic gram-negative bacteria. To obtain an amidase with impact(s) on gram-negative bacteria as well as gram-positive bacteria, in this study, MurNAc-LAA from Bacillus subtilis (B. subtilis)-bacteriophage Siphoviridae was heterologously expressed in Escherichia coli (E. coli). The antibacterial properties of the endolysin were investigated in relation to its secondary and tertiary structure.
Materials and Methods :
Muramyl- N-Acetyl -L-Alanine Amidase (LysM2) gene construction: The gene and protein sequences of MurNAc-LAA from the Bacillus phage SPP1 in B. subtilis (Accession number: X97918) were obtained from the Virus-Host database (GenBank number CAA66518, NCBI). The GC content was adapted for expression in E. coli by GenScript codon optimization software (http://www. genscript.com/). The optimized MurNAc-LAA gene in our laboratory, MurNAc-LAA-MMI (LysM2, appendix 1), was synthesized by Gene Transfer Pioneer company (GTP, Iran) and cloned into pUC57.
In silico study of LysM2 protein: Using NCBI Conserved Domain Database (CDD) 12 and InterPro database, the functional domain of LysM2 was investigated 13. Tree Viewer v 1.17.4 software was used to generate a MurNAc-LAA cladogram containing the LysM2 endolysin 14.
RNA secondary structure was used to study RNA stability 15. The molecular weight of LysM2 protein, the abundance of amino acids, and half-life of protein were determined by ProtParam software (https://web. expasy.org/ protparam/). The hydrophobicity of LysM2 was plotted on the Kyte-Doolittle scale using the ProtScale tool 16. Secondary structure was obtained by the self-optimized prediction method (SOPMA) (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page/NPSA/npsa sopma.html) 17 and PSIPRED server (http://bioinf.cs.ucl.ac.uk/psipred/). The transmembrane helix was predicted using TMHMM v 2.0 18. The three-dimensional (3D) model for the amidase was predicted using Protein Homology/analogY Recognition Engine V 2.0 (Phyre2) software 19 and validated by PROSA-web (Protein Structure Analysis) 20. The 3-D structure of molecules was analyzed by the Ramachandran plot 21. FT site server (https://ftsite. bu.edu/) predicted the active-site (s) in the tertiary structure.
Localization of LysM2 protein in the host cell and antigenicity: The subcellular location of LysM2 protein in the host cell was predicted by the Virus-mPLoc software (https://www.psort.org). The antigenicity of LysM2 protein was evaluated by the Vaxijen software 2 (http://www.ddgpharmfac.net/vaxijen/VaxiJen/VaxiJen.html) 22. The potential for toxicity of LysM2 was predicted using the BTXpred (Prediction of Bacterial Toxins) server (http://crdd.osdd.net/raghava/btxpred/).
Cloning and expression of LysM2 gene: The cutting sites for BamHI (Sinaclon, Iran) and SacI (Termo Fisher Scientific, USA) restriction enzymes were respectively introduced at 5′ and 3′ end of the coding sequence. The LysM2 gene was restricted using BamHI/SacI from pUC57/LysM2, and ligated into pET28a vector with the same enzymes. The generated construct, pMMI2 (pET28a/LysM2), was introduced into the E. coli B121 (DE3) cells. The transformed cells were selected on LB agar supplemented with 40 μg/ml kanamycin (Sigma-Aldrich, USA). The cloning procedure was confirmed by BamHI/SacI digestion analysis.
The expression of pMMI2 was induced by 1 mM isopropylthio-β-galactoside (IPTG) (DNAbiotech, Iran) at 28°C for 20 hr. The cells were suspended in 20 mM NaH2PO4, 500 mM NaCl, and 50 mM Imidazole (Merck, Germany) buffer (pH=7.2-7.4), sonicated on ice for 10 min (90 w, 30 s at 7 s interval), and centrifuged at 15000×rpm for 20 min. The supernatant was collected. As a negative control, all the above steps were carried out for an uninduced E. coli BL21(DE3) harboring pMMI2. Fractions were analyzed on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) for protein expression of pMMI2. Protein purification was done by NI-NTA affinity chromatography (DNAbiotech, Iran). The column was eluted with a range of Imidazole concentrations as 20-150 mM (50 mM NaH2PO4; 300 mM NaCl; pH=7.7), dialyzed against Phosphate-Buffered Saline (PBS), and analyzed on SDS-PAGE. The amount of LysM2 protein was measured by Bradford assay.
Evaluation of the enzyme activity: The lysed cell wall of the indicator bacteria, Streptococcus pyogenes (S. pyogenes) (ATCC 19615), Bacillus subtilis (B. subtilis) (ATCC 12711), Bacillus )sp) (SUBC1001), Staphylococcus aureus (S. aureus) (ATCC25923), Pseudomonas aeruginosa (P. aeruginosa) (ATCC27853), E. coli (ATCC25922), Salmonella )sp.( (SUBC1002), Proteus )sp.( (SUBC1003), Klebsiella pneumonia (K. pneumonia) (ATCC13883), Salmonella typhimurium (S. typhimurium) (SUBC1005), Proteus vulgaris (P. vulgaris) (SUBC1006), and a clinical K. pneumoniae (SUBC1004) was prepared for evaluating the enzyme activity as previously described 23. Briefly, the heat-killed indicator bacteria were added to LB medium. The plates were punched to make 8 mm wells followed by adding 100 µl of the purified enzyme. The strains with the clear zone were selected for evaluating their antibacterial activities. The Activity Unit (AU) of the endolysin was calculated as previously described (p<0.05) 24:
Endolysin activity (mm2/ml) = (Lz – Ls)/V
Lz = Clear zone area (mm)2, Ls = Well area (mm)2, V = Volume of sample (ml)
The boiled-culture of the indicator bacteria with OD600 of 0.6 was subjected to 1:2 serial dilution of the endolysin for 40 min at 25°C. A dilution of the enzyme that reduced the turbidity of the heat-killed indicators to 50% was introduced as the activity of LysM2 enzyme.
Next, 500 µl of boiled-culture of the indicator bacteria with OD600 of 0.6 was resuspended in the volume of 20 mM Tris HCl (pH=7.5) and 500 µl of the enzyme was mixed in a corvette, and the buffer without endolysins was used as a negative control. The mixtures were incubated at 37°C for 70 min. A volume of the enzyme that reduced the turbidity of the heat-killed indicators to 50% was introduced as the enzyme activity.
Results :
LysM2 gene construction : The LysM2 gene with 828 nucleotides in length was subcloned from pUC57 in pET28a. The codon preference was adjusted for E. coli and the conservation of the LysM2 sequence was evaluated by GenScript software.
In silico study of LysM2 protein: Sequence analysis: The most similar protein to MurNAc-LAA from B. subtilis phage was found in Firmicutes bacteria upon a comparison among 100 endolysins harboring the MurNAc-LAA domain in the GenBank (Figure 1A). BLASTp homology analyses revealed a conservativity (86% to 96% identity) among MurNAc-LAA enzymes from Bacillus phages. The catalytic domain of LysM2 protein against the bacterial cell wall was investigated in silico (Figure 1B). The whole gene similarity was 80% at nucleotide level within the MurNAc-LAA family. The prediction of the mRNA structure for LysM2 confirmed the availability of enough free energy for the optimum expression.
The study of physico‑chemical parameters: ProtParam analyses demonstrated that LysM2 is a protein of 271 amino acids in length with the alpha-index of 83.87, half-life of more than 10 hr in E. coli, 20 hr in yeast, and 30 hr in mammalian reticulocytes (in vitro), and the molecular weight of 29.7 kDa. The LysM2 protein showed a stability index of 40.48 where a value of more than 20.48 indicates stability in protein structure. Taken together, the results displayed that LysM2 is a stable protein in structure.
Evaluation of the secondary and tertiary structure: The secondary structure of the LysM2 enzyme exhibited a proportion of 34.2% of alpha-helix, 23.23% of extended strand, 4.43% of beta-turn, and 36.9% of random coil (Figures 2A and 2B). A total residue of 248 (91.5%) was modeled with 100.0% confidence in the crystal structure of the MurNAc-LAA from E. coli. The frequency of positively charged amino acids is higher than the N-terminal end of the protein (aa 1-150) (Figure 2C). LysM2 was found as a hydrophobic nonpolar molecule due to the presence of hydrophobic residues at the N-terminal end. The PI value was detected as 9.67 for the lysine and alanine. These two amino acids have tendency to form alpha helices. The active sites for LysM2 enzyme are demonstrated in figure 2D. The FT software predicted three active sites in the tertiary structure of the LysM2 enzyme. The second active site was similar to the active sites predicted by NCBI-tool. Using RAMPAGE server, the proposed model for LysM2 protein was evaluated. The results showed that 2.2% of the residues were in the outlier region, 5.6% in the allowed area, and 92.1% of amino acids in the favored region. Consequently, 97.7% of the amino acids were located in a stable situation. Z-score was measured as -6.12 in comparison with the pre-determining structures (Figures 3A and 3B).
Localization of LysM2 protein in the host cell and the antigenicity: LysM2 protein was predicted to localize in the protoplasm of the host cells. The antigenic properties were analyzed using Vaxijen software 2. The value of 0.4, lower than 0.5038, suggests no antigenic properties for LysM2. The BTXpred server predicted the toxicity of LysM2 protein for bacteria based on analyzing the primary structure (SVM model).
Cloning and expression of LysM2 gene: The construct of LysM2 gene (pMMI2) was transformed into E. coli BL21(DE3). The purified protein was run on a 12% polyacrylamide SDS-PAGE. The protein of 33 kDa related to LysM2 was detected on the SDS- PAGE (Figure 4). The yield of protein from E. coli was obtained as 0.8 mg protein per 1 gr cell and the specific activity was 2 U per 0.2 mg/ml protein.
Antibacterial activity assay: The enzyme activity was measured by monitoring the cell wall lysis via agar well-diffusion method. The results of the agar diffusion assay showed a relatively optimal antimicrobial activity of 2950 AU in diameter against K. pneumoniae, 6010 AU for S. typhimurium, and 6730 AU for P. vulgaris (Figure 5A).
The culture turbidity of the indicator strains was reduced subject to the natural extraction of the LysM2 protein. The lytic activity was determined only on the Proteus sp. as depicted in figure 5B. The unit activity was calculated as 2 Unit/ml.
Discussion :
Endolysins lyse the cell wall of gram-positive bacteria, while an outer membrane protein supports gram-negative bacteria. However, endolysins either in companion with holins or with a cationic N or C-terminal tail kill the gram-negative bacteria 7.
In this study, an endolysin called LysM2, from a Siphoviridae phage was exogenously expressed in E. coli. The 29.7 kDa amidase was analyzed in silico. The presence of 34.2% alpha-helix with positive hydrophobic amino acids in the N-terminal end of the enzyme confirmed LysM2 as a nonpolar molecule similar to lipopolysaccharide molecules in the membrane. These data suggest the possible interaction(s) between the N-terminal region and lipopolysaccharide in the outer membrane of the bacteria 25.
Amphipathic peptides with a modular structure containing alpha-helix kill the bacteria by a mechanism independent of their enzyme activity. Positively charged amino acid in companion with amphipathic helical structure showed a stronger bactericidal activity 26. MurNAc-LAA contains amino acids with a net positive charge (Lysine residues) at C-terminus which help them to interact with the inner layer of the cytoplasmic membrane. In this study, the 33 kDa enzyme is featured as a nonpolar protein with the hydrophobic N-terminal structure in alpha-helix similar to lipopolysaccharide of gram-negatives outer membrane. LysM2 contains a lysine rich C-terminus similar to other members of MurNAc- LAA family.
Regamey and Karamata cloned and expressed blyA gene encoding 39.6 kDa MurNAc-LAA. Using mutagenesis method to determine the function of blyA, the role of this enzyme in Spβ phage-mediated cell-lysis was confirmed. Furthermore, two ORFs encoding polypeptides as holins are located at the downstream of blyA gene and they are involved in releasing endolysin in the gram-negative bacteria 27. Lai et al identified a phage lysozyme, LysAB2. Antibacterial analysis demonstrated the impact of LysAB2 on the cell wall of A. baumannii and S. aureus. The recombinant LysAB2 lysed cell wall of several gram-negative and gram-positive bacteria 28.
The membrane destabilizing agents such as poly-l-lysine, polymyxin B, ethylenediaminetetraacetic acid disodium (EDTA), or highly charged-hydrophobic amino acid residues increase the membrane permeability to lysins 29. Oliveira et al studied the endolysins from Salmonella phage including Lys68, SPN1S, and SPN9CC with anti-gram-negative and without anti-gram-positive activities. A Salmonella phage endolysin (Lys68) in combination with EDTA, citric acid, and malic acid lyse many gram-negative bacteria. The enzyme caused 3 to 5 log reductions in bacterial load/ CFUs after 2 hr against S. typhimurium LT2 and diminished stationary-phase and bacterial biofilms by about 1 log 30. This evidence demonstrates that in the absence of holins, a membrane destabilizer like EDTA is required for cell wall destruction. In contrast, there are some documents showing no need for EDTA or holins to make the membrane permeable. Dong et al produced exogenous Maltocin P28, a phage-tail like bacteriocin, that harbors the conserved domain of lysozyme-like superfamily. The recombinant P28 destroys gram-negative bacteria in the absence of EDTA. A putative helix domain in the N-terminal hydrophobic region was documented for P28 similar to LysM226.
To count the endolysins as antibacterial agents, the mechanism of action for these enzymes must be determined. Investigation on the mechanism of action of the peptidoglycan cleaving enzyme was done by Yakhnina and Bernhardt. They demonstrated that the multiprotein Tol-Pal system in the envelope of gram-negative bacteria is linked with remodeling of peptidoglycan in outer membrane of E. coli and with the hypersensitivity to many antibiotics. E. coli Tol-Pal mutants have incomplete peptidoglycan layer 31.
Heidrich reported that in the presence of antibiotics, three amidases are involved in splitting murein septum during cell division in E. coli. The murein cross-bridges cleaves and blocks cell division 32. Some studies revealed that the engineering of catalytic domain in MurNAc-LAA amidase presumably is not a short way to acquire the desired endolysins. Morita et al showed that both the C-terminal cell-binding and the N-terminal enzymatic domains are required for the enzymatic activity of phage endolysin from Bacillus amyloliquefaciens. This protein has two helical peptides at the C-terminus of endolysin that may bind to the lipopolysaccharide of P. aeruginosa PAO16.
Exogenous expression of amidase in E. coli is a promising method for the enzyme production. In this study, an E. coli optimized amidase, LysM2, showed anti-Proteus (sp), a relatively anti-K. pneumoniae, anti-S. typhimurium, and anti-P. vulgaris activities. Scheurwater et al studied, cloned, and expressed the N-acetylmouramoyl-L-alanine amidase gene B (AmiB) from P. aeruginosa in E. coli. AmiB (50 kDa) digests M. luteus peptidoglycan as a substrate in 5 mM sodium phosphate buffer at pH=6.5. The specific activity of 4.4 ∆OD.660U-min -1 mg protein-1 was reported for AmiB. Moreover, it degrades the purified insoluble peptidoglycan from gram-negative bacterium, P. aeruginosa 1.
Magdalena Plotka et al showed that the antibacterial spectrum of endolysins is related to the type and structure of the outer membrane. Acinetobacter and Pseudomonas were susceptible to Ts2631 endolysin. The high number of phosphate groups per lipopolysaccharide molecule in the cell wall of these two bacteria help the Ts2631 with cationic N-terminal for destabilizing activity. In contrast, Enterobacteria lacks such a highly negative charge per lipopolysaccharide molecule, protecting against Ts2631 function 33,34. Recently, a lytic enzyme, LysC, from Clostridium intestinale was introduced. Its antibacterial activity is mediated by positively charged N-terminal extension 35.
Haddad et al designed a chimeric CHAP-amidase for improving the stability and solubility of the enzyme. Significant antibacterial activity against S. aureus MRSA252 was reported applying 1 µg/ml chimeric CHAP-amidase. The protein showed strong antibacterial activity against S. epidermidis, S. aureus, and Enterococcus (sp) while notable antibacterial activity was detected against E. coli and L. lactis. The synergistic effect of CHAP-amidase and vancomycin indicated an 8-fold decrease in the minimum inhibitory concentration of vancomycin 31.
The MurNAc-LAA belongs to either amidase-2 or amidase-3 zinc-dependent families, cleaves the amide linkage between N-acetylmuramic acid and L-alanine. The amidase-2 domain is formed by a water molecule with the side chains of histidine, cysteine residues, and a zinc ion. The catalytic domain in PlyPSA and CD27L proteins belongs to amidase-3 family, showing a different enzymatic activity despite highly similar mode of action and 3D structure 10.
Furthermore, other documents demonstrate that catalytic modular domain of lysins is efficiently substituted with other lysins for the new bactericidal features 29. In this study, the enzyme with a catalytic domain of endolysin, LysM2, inhibited P. vulgaris, K. pneumoniae, and S. typhimurium. Both cell binding domain and catalytic domain contribute in the lytic activity of MurNAc-LAA amidases. The cell wall binding domain in MurNAc-LAA may alter the toxicity of the endolysin via increasing or decreasing the rate of enzyme-substrate contact. Therefore, engineering the binding domain in the amidase can optimize its lytic effects on the targeted bacteria. Engineering the binding domain and catalytic domain of LysM2 should be studied in further research.
Conclusion :
The gene and amino acid sequences of peptidoglycan hydrolase enzymes in lysogenic phages of the Siphoviridae family hosted in B. subtilis were investigated in this study. The molecular weight of the protein was calculated as 33 kDa with 271 amino acids in length. The enzyme half-life was estimated to be at least 10 hr in E. coli. The protein secondary structure predominantly contains alpha-helix. LysM2 exhibited no antigenic properties in in silico analyses. LysM2 protein showed an antimicrobial activity against K. pneumoniae, and S. typhimurium in agar diffusion assay and P. vulgaris and Proteus (sp) in turbidity assay. Taken together, our results confirmed that MurNAc- LAAn-MMI is an inhibiting agent for gram-negative bacteria.
Acknowledgement :
This study was fully funded by Iran National Science Foundation, INSF NO. 95828352, Tehran. The authors acknowledge the assistance of the staff at Deputy for Research and Technology, INSF, and Semnan University, Semnan, Iran for supporting this study.
Conflict of Interest :
The authors declare that they have no competing interest.
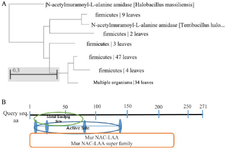
Figure 1. A) The phylogenetic tree for the optimized LysM2, based on the Neighbor-Joining method. B) In silico investigation of the catalytic domain of LysM2
|
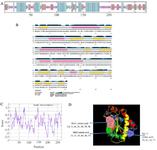
Figure 2. A, B) Secondary structure prediction for LysM2 protein. C) Frequency of positively charged amino acids is higher in the N-terminal protein. D) The three-dimensional structure of LysM2 protein. Purple, red, and blue colors indicate extended strand, coil, and helix, respectively in both figures, A and D.
|

Figure 3. A) Evaluation of LysM2 protein stability based on Ramachandran plot showed 2.2% of the residues were in the outlier region; 5.6% in the allowed area, and 92.1% of amino acids in the favored region. B) The results of analyzing LysM2 protein with the ProSA web server; Z-score plot, NMR spectroscopy (dark blue), and X-ray crystallography (light blue); the plot showed local model quality using plotting energies as a function of amino acids position in the sequence.
|
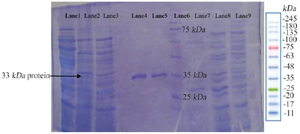
Figure 4. The photogram of LysM2 protein on 12% SDS-PAGE. The relevant 33 kDa band is indicated in the Lanes 1, 2: LysM2 protein, Lane 3: Control, Lanes 4, 5: Purified protein, Lane 6: Ladder protein 10-250 kDa, Lane 7: Purified control without IPTG induction, Lane 8: E. coli BL21DE3, Lane 9: Pre induction of pET28a without the insert.
|

Figure 5. A) Agar diffusion of LysM2 against Proteus (sp). B) The lytic activity of LysM2 against Proteus (sp).
|
|