Association of CALHM1 Gene Polymorphism with Late Onset Alzheimer’s Disease in Iranian Population
-
Jafari Aqdam, Meysam
-
Genetic Research Centre, University of Social Welfare and Rehabilitation Sciences , Tehran, Iran
-
Kamali, Koorosh
-
Reproductive Biotechnology Research Centre, Avicenna Research Institute, ACECR , Tehran, Iran
-
Rahgozar, Mehdi
-
Epidemiology and Biostatistics Department, University of Social Welfare and Rehabilitation Sciences , Tehran, Iran
-
Ohadi, Mina
-
Genetic Research Centre, University of Social Welfare and Rehabilitation Sciences , Tehran, Iran
-
Manoochehri, Mehdi
-
Genetic Research Centre, University of Social Welfare and Rehabilitation Sciences , Tehran, Iran
-
Tahami, Ali
-
Reproductive Biotechnology Research Centre, Avicenna Research Institute, ACECR , Tehran, Iran
-
Bostanshirin, Leila
-
Genetic Research Centre, University of Social Welfare and Rehabilitation Sciences , Tehran, Iran
-
 Khorram Khorshid, Hamid Reza
Hamid Reza Khorram Khorshid, M.D., Ph.D., Genetic Research Centre, University of Social Welfare and Rehabilitation Sciences, Tehran, Iran, Tel: +98 21 22180138 E-mail:hrkk1@uswr.ac.ir
Khorram Khorshid, Hamid Reza
Hamid Reza Khorram Khorshid, M.D., Ph.D., Genetic Research Centre, University of Social Welfare and Rehabilitation Sciences, Tehran, Iran, Tel: +98 21 22180138 E-mail:hrkk1@uswr.ac.ir
-
Genetic Research Centre, University of Social Welfare and Rehabilitation Sciences , Tehran, Iran
-
Reproductive Biotechnology Research Centre, Avicenna Research Institute, ACECR , Tehran, Iran
Abstract: Alzheimer's disease (AD) is a genetically heterogeneous neurodegenerative disease and Late-Onset type (LOAD) is the most common form of dementia affecting people over 65 years old. CALHM1 (P86L) encodes a transmembrane glycoprotein that controls cytosolic Ca2+ concentrations and Aß levels and P86L polymorphism in this gene is significantly associated with LOAD in independent case controls in a number of studies. This study was performed to determine whether this polymorphism contributes to the risk for LOAD in Iranian population. One hundred and forty one AD patients and 141 healthy controls were recruited in this study. After extraction of genomic DNA, the genotype and allele frequencies were determined in case and control subjects using PCR/RFLP method. The statistical analysis showed a significant difference in the heterozygote genotype frequency in case and control groups and polymorphic allele had a protective role between two groups. Also after stratifying the subjects by their APOE-e4 status, no significant association was observed. Our study suggests that P86L polymorphism could be a protective factor for late-onset Alzheimer's disease (LOAD) in Iranian population. However, to confirm these results, further study with a bigger sample size may be required.
Introduction :
Alzheimer's disease (AD) is a progressive neurodegenerative disorder characterized by a massive loss of neurons in different brain regions and by the presence of cerebral senile plaques comprised of aggregated Aß peptides (1). Late-Onset Alzheimer's Disease (LOAD) is a type of AD and the most common form of dementia affecting people over 65 years old (2). Because of its strong genetic heterogeneity, the etiology of the disease is complex.
Materials and Methods :
To determine the possible role of P86L polymorphism in CALHM1 gene in developing LOAD in our population, a case-control study was performed. A sample of 141 AD patients (mean age 77.82 yr) and 141 healthy controls (mean age 78.26 yr) (from 7 old people’s home throughout Tehran with various genetic backgrounds) were recruited for the study.
The diagnosis of Alzheimer’s disease in patients was confirmed by qualified psychiatrists according to the DSM-IV criteria and control subjects were selected through the assessment of their medical histories and physical conditions. The recruited subjects in control group did not have any known neuropsychiatric disorders. We assumed genetic background, gender, sex, age, and education as potential confounders. After obtaining informed consent from participants or their families, the information regarding the age, sex, ethnicity, job and education were asked and recorded.
Finally 5 cc of peripheral blood samples were collected in tubes containing 200 µl of 0.5 M EDTA. Genomic DNA was extracted from peripheral blood using the salting-out method and a pair of primers (Forward: 5`-CCTGGTCATGAACAACAACG-3` and Reverse 5`-AGGCACAGAGGAAGCATTT G-3`) was used to amplify the relevant fragment and analyze P86L polymorphism (appropriate primers were designed using Primer3 program). Genotyping of samples determined using PCR amplification, restriction digestion and Polyacryl Amide Gel Electrophoresis (PAGE). To amplify the 182 bp fragment, PCR was performed with 1.5 mM MgCl2 and 60 °C annealing temperature. The 182 bp PCR products were digested using HpaII restriction enzyme (10 U/µl, Fermentas). The product of digestion loaded on 8.5% polyacryl amide gel for electerophoresis. Digestion using this enzyme produced 53, 8 and 121 bp fragments for wild type and 61, 121 for mutant.
The Chi square (?2) or Fisher’s exact test with Open Epi Version 2.2 (free statistical software) was performed to compare genotype and allele frequencies in the study groups. CC genotype and C allele were assumed as reference group for statistical analysis. To assess the role of interaction of APOE e4 allele and sex with this polymorphism, the stratified analysis was performed regarding the existing APOE e4 PCR results with these samples. The different allele and genotype frequencies of APOE gene were determined in our previous study using PCR/ RFLP method (8).
Result :
Total 282 DNA samples were analyzed using the PCR and Restriction Fragment Length Polymorphism (RFLP) method. The samples consisted of 141 AD patients and 141 healthy controls. The distribution of important potential confounders in the studies groups were similar (Table 1). The distribution of P86L genotype and allele frequencies are summarized in table 2. The statistical analysis of the patient’s genotype and allele frequencies showed significant difference in heterozygote genotype (CT) and T allele (mutant) had a protective role between two case and control groups.
We performed the stratified analysis (Fisher’s exact test) by APOE e4 allele and sex for P86L genotype and alleles (Tables 3 and 4). The results for male sex and e4 Allele positive were not statistically significant and for female sex and e4 Allele negative, the result did not change with the crude data. It can be concluded that we could not find the effect modifier role for variables sex and e4 allele on CALHM1 genotypes and alleles between case and control groups.
Discussion :
Existence of linkage for chromosome 10 with Alzheimer disease had been reported repeatedly and CALHM1 gene on this chromosome is one of the candidate genes for this association. This gene encodes for integral membrane protein that acts as a calcium channel. Calcium pathway has a fundamental role in the pathogenesis of AD and this channel is important in this way by controlling homeostasis of calcium ion. It had been shown in previous studies that rs2986017 SNP in CALHM1 gene (P86L) is associated with both an increase risk for LOAD and a significant dysregulation of Ca2+ homeostasis and APP metabolism (1, 9).
In the present study 282 participants (141 AD and 141 controls) were recruited. After carrying out PCR and RFLP techniques, data were used for statistical analysis. Analysis of acquired data with statistical methods showed that T allele (mutant) has a protective role and is nearly significant between cases and controls (p= 0.057).
Also comparing the frequency of genotypes showed significant difference in distribution of heterozygote genotype in two groups (p= 0.004). But the interaction analysis of CALHM1 genotype with APOE e4 allele did not indicate any significant difference.
The result of our study in Iranian population is in agreement with some studies in other parts of the world. In a study carried out in the USA with sample size comprising 2043 cases and 1361 controls, a significant difference was shown (1). Another study was done with the sample size consisting of 2500 cases and controls in Japan (2009) and the difference between this polymorphism and Alzheimer disease was not significant (10).
In a study carried out in China (2009) the number of 198 Alzheimer patients was examined and a significant difference was shown (11). In another study carried out in Spain the difference between this polymorphism with AD was significant (12). In study carried out in the USA (2009) with sample size including 510 cases and 524 controls, no significant difference was shown (13). Therefore, the findings of our study in the Iranian population is supported by certain studies in the world (1, 11, 12) and is in contrast with the results of other studies carried out in this subject in other parts of the world (10, 13 - 17).
There is a possibility that P86L polymorphism could influence the appearance onset of signs and symptoms (delay or postpone the beginning of disease) in affected individuals. Because in this study we did not have the onset age of the dementia symptoms, therefore we could not identify the role of this polymorphism in the onset of signs and symptoms of the Alzheimer’s disease.
In conclusion, our results indicate significant difference in heterozygote genotype distribution between AD patients and controls, but a larger sample size may be needed for stronger confirmation of our findings.
Acknowledgement :
We express our sincere thanks and gratitude to all Alzheimer's and control persons or their families for their kindly participation in this study. We also thank Iran Alzheimer Association for their sincere collaborations.

Table 1. Comparison of mean age, sex and education levels between AD case and control subjects using t-test and χ2 test analysis
|
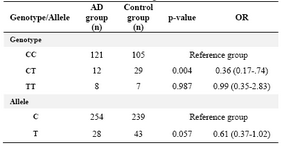
Table 2. The distribution of P86L genotype and allele frequencies in CALHM1 gene
|
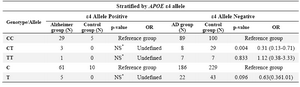
Table 3. Stratified analysis of CALHM1 genotypes and alleles by ε4 allele
NS*: Not statistically significant by Fisher's exact test
|
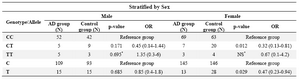
Table 4. Stratified analysis of CALHM1 genotypes and alleles by sex
NS*: Not statistically significant using Fisher�s exact test
|
|