Expression and Characterization of Two DNA Constructs Derived from HIV-1-vif in Escherichia coli and Mammalian Cells
-
Zamani, Fatemeh
-
Department of Hepatitis, AIDS and Blood Borne Diseases, Pasteur Institute of Iran, Tehran, Iran
-
Department of Biology, Science and Research Branch, Islamic Azad University, Tehran, Iran
-
Bolhassani, Azam
-
Department of Hepatitis, AIDS and Blood Borne Diseases, Pasteur Institute of Iran, Tehran, Iran
-
Shahbazi , Sepideh
-
Department of Hepatitis, AIDS and Blood Borne Diseases, Pasteur Institute of Iran, Tehran, Iran
-
Faghih, Ahmad
-
Department of Hepatitis, AIDS and Blood Borne Diseases, Pasteur Institute of Iran, Tehran, Iran
-
Department of Biology, Science and Research Branch, Islamic Azad University, Tehran, Iran
-
 Sadat, Seyed Mehdi
Department of Hepatitis, AIDS and Blood Borne Diseases, Pasteur Institute of Iran, Tehran, Iran, Tel/Fax: +98 21 66969291; Email:mehdi_sadat@pasteur.ac.ir
Sadat, Seyed Mehdi
Department of Hepatitis, AIDS and Blood Borne Diseases, Pasteur Institute of Iran, Tehran, Iran, Tel/Fax: +98 21 66969291; Email:mehdi_sadat@pasteur.ac.ir
Abstract: Background: Acquired immunodeficiency syndrome (HIV/AIDS) is still a major global concern and no effective therapeutic vaccine has been produced to prevent the problem. Among HIV-1 proteins, vif as a basic cytoplasmic protein of HIV-1 is involved in late stages of viral generation and plays important role in HIV-1 virion replication. It also increases the stability of virion cores, which probably inhibits early degradation of viral entry. Therefore, it seems rational to apply this protein as a vaccine based on its impact on HIV-1 life cycle. This study aimed at cloning, expression and production of vif protein as an HIV-1 vaccine candidate.
Methods: In this study, vif sequence was amplified from pLN4-3 plasmid including HIV-1 vif gene and then cloned in pET23a to generate the recombinant plasmids of pET23a/vif with hexahistidine tags. BL21 competent cells were transformed to obtain the protein of interest. Ni-NTA column was used to purify the protein of interest and western blotting confirmed vif protein using anti-His tag antibody. In order to express the gene of interest in eukaryotic cells, vif was sub-cloned into pEGFP plasmids and HEK 293-T cells were transfected. Flow cytometry was then applied to evaluate GFP expression.
Results: vif protein was expressed in BL21)DE3) strain and identified as a23 kDa band in SDS-PAGE and confirmed by anti-His antibody in western blotting. The purified protein concentration was 173.3 μg/ml using Bradford assay. HEK-293T cells were successfully transfected by recombinant pEGFP plasmids and flow cytometry confirmed the cell transfection.
Conclusion: vif protein can be expressed in mammalian cells and may be a proper protein subunit vaccine candidate against HIV-1.
Introduction :
The Human Immunodeficiency Virus type 1 (HIV-1) has been the causative agent of AIDS for nearly 40 years. It has infected 74.9 million individuals since its outbreak. It is estimated that there were 37.9 million people living with HIV in 2018 despite the great effort done to overcome it 1,2. HIV is now considered a chronic illness which is manageable due to great preventive strategies offered by Antiretroviral Therapy (ART) 3. Despite this advance in virus life cycle, ART is not capable to either eradicate the virus or cure the infected people. The virus mostly remains in multiple organs of reservoir host although viral load is not detectable in the host blood 4-6. In order to approach a therapeutic vaccine, many strategies have been applied
along with preventive studies to defeat HIV challenges through identification of virus characteristics 7-10.
Apart from main genes, HIV genome includes some accessory genes (rev, tat, nef, vpu, vpr, and vif) which were initially considered unessential in infection. Nevertheless, it has been shown that they are important for severe infection in vivo. New evidence shows that certain viral proteins, like vif, are capable to evade host antiviral innate immune responses 11-13. vif is a basic cytoplasmic protein of HIV-1 which is involved in late stages of viral replication and participates in generating infectious HIV-1 virions along targeting CEM/15 APOBEC3G protein. The virus is unable to replicate in non-dividing cells such as Peripheral Blood Mononuclear Cells (PBMCs), macrophages, and H9 cells in absence of vif. This protein is better preserved, in comparison with other HIV genes, in closely related strains such as gag and env 14-16. Small amount of vif was found to be packaged into recently synthesized progeny, but the validity of this issue is not entirely clear. vif mutant HIV-1 virions show structural deformities within the cone-shaped core. Moreover, vif enhances the stability of virion cores, which may inhibit early degradation upon viral entry 17-19. The influence of vif on non-permissive cells, which is critical for proviral DNA synthesis, is extremely dependent on cellular factors associated with vif 11,20.
Therefore, according to the importance of vif in the HIV life cycle, applying it as an HIV vaccine candidate seems to be logical. Doi et al investigated SA1D2prox and vif gene by chimeric approaches applying HIV-1 subtypes B and C. The generated recombinant clones were carrying chimeric sequences of SA1D2prox and vif within the vif coding region 21. In this study, HIV vif gene cloning and expression in bacterial platform was investigated using pET23a expression vector. Also, vif gene was applied in the mammalian cell using pEGFP-N1 to evaluate its expression in both prokaryotic and eukaryotic systems.
Materials and Methods :
Bacterial strains and plasmids: DH5α and BL21(DE3) strains of Escherichia coli (E. coli) were used for cloning and protein expression. The prokaryotic and eukaryotic expression vectors were pET23a (+), pEGFP-N1, pNL4-3, and plasmid including HIV-1 vif gene; plasmids were gifts from Hepatitis, AIDS and Blood Borne Diseases Department, Pasteur Institute of Iran.
vif gene amplification and cloning: PCR technique using Taq DNA polymerase (Roche Applied Science, Germany) including a specific primers of vif F: 5’- GAGCTAGCGGTACCATGGAA AACAGATG -3’, vif R: 5’- CTAAAAAAGCTTGTG TCCGTTCATTGTATGGCTCCC -3’ containing a restriction enzyme site (Shown in bold) for Nhel in the forward primer and a Hindlll restriction in the reverse primer was applied to amplify vif gene. Steps for PCR included initial denaturation at 95°C for 2 min following by 30 cycles of denaturation at 95°C for 30 s, annealing at 61°C for 45 s, extension at 72°C, and final extension at 72°C for 5 min. The amplified products were then observed on 1% agarose-gel electrophoresis. The PCR product of the expected size was purified using PCR product purification kit (Qiagen, Germany) according to the manufacturer’s instruction. The PCR product was digested with Nhe l/Hind lll and ligated to the linearized pET 23a to generate the recombinant plasmids pET 23a/vif with hexahistidine tags. Finally, the cloned pET23a/vif vector was transformed into DH5α competent E. coli cells using the heat shock method. In order to confirm the presence of inserted gene in PET 23a vector, restriction enzyme analysis was applied. Finally, the reading frame of target gene was confirmed by sequencing.
Protein expression, confirmation, and purification: E. coli BL21(DE3) was transformed with recombinant pET23/vif. Protein expression was induced by IPTG (Isopropyl β-D-1-thiogalactopyranoside) addition (Sigma, USA) to mid-log phase culture of bacteria at 37°C. The cell pellets were harvested and analyzed by SDS-PAGE. Finally, the bacterial lysate was transferred to nitrocellulose membranes (Sigma, USA) according to the general procedure described by Towbin et al 22. Then, the membrane was stained with Ponceau to visualize the proteins and blocked by overnight incubation at 4°C with BSA blocking buffer (10 ml of TBS1X+2/5 g of BSA (Sigma, USA) +100 μl of Tween 20). Anti-His Tag conjugated antibody (Abcam, USA) was applied to confirm the protein of interest. The expressed protein was purified by Ni-NTA sepharose-based affinity chromatography under naturing conditions. Briefly, lysis buffer containing 5 mM of imidazole was used to solubilize the cell pellet and after 30 min incubation and sonication, the lysate was applied to the Ni-NTA column following three times of washing steps. Next, 250 mM of imidazole elution buffer was used to purify the protein. Finally, high concentration of imidazole was exchanged by dialysis bag (Thermo Fisher Scientific, USA) to PBS buffer and dialyzed overnight at 4°C. The purified protein concentration was calculated by BCA protein assay kit (Pierce, USA) through measurement of optical density at 280 nm 23.
Eukaryotic expression vector: The pEGFP-vif expression vector was constructed by sub-cloning the vif gene into the Nhel/HindIII site of the related vector. The DNA construct containing vif gene (pEGFP-vif) was purified at a large scale using Midi-kit (Qiagen, USA). The presence of the inserted vif fragment was confirmed by colony PCR as detected on gel electrophoresis.
vif gene transfection and quantitation: Human HEK-293T cells were seeded in RPMI 1640 Complete Medium (Sigma, USA) containing 10% heat inactivated fetal calf serum (Gibco, Germany) at 37°C. Transient transfection in HEK-293T cells with pEGFP-vif (vif expression vector) and pEGFP-N1 (Positive control) was accomplished using TurboFect Transfection Reagent in eukaryotic cells (Fermentas, Germany). The negative controls were HEK-293T cells.
For transfection, 2 μg of plasmid was pre-incubated with 4 μl of TurboFect in a total volume of 25 μl and incubated at room temperature for 20 min to permit the formation of DNA/ TurboFect complexes. The complexes were then added to 2×105 HEK-293T cells in each well using a 12-well plate. The cells were harvested 24 hr after transfection, washed, and resuspended in PBS. Finally, flow cytometry analysis (Partec, Germany) was used to determine the number of fluorescent cells expressing vif-GFP and the quality of DNA expression was also evaluated using fluorescence microscopy.
Results :
Generation of pET23a/vif and pEGFP-N1/vif plasmids: The plasmids with T7 and CMV promoters (pET 23a and EGFP-N1, respectively) were applied which produced periplasmic C-terminal His tagged and EGFP tagged vif proteins. Nhel and Hindlll restriction enzymes were used to digest the gene of interest which was inserted into the pET 23a/vif and pEGFP-N1/vif and confirmed by PCR and restriction analysis. The results showed a sharp band of 570 bp for vif gene in 0.1% agarose gel (Figure 1). Moreover, the plasmid constructs were verified by sequencing.
Expression and purification in E. coli BL21 (DE3): The recombinant vif protein was expressed in BL21)DE3) strain. The vif protein migrated as a clear band of ~23 kDa in SDS-PAGE which was identified using anti-His antibody in western blotting (Figure 2). An optimal expression condition for protein was obtained following overnight incubation after induction with 2.5 mM of IPTG until OD600 nm reached 0.4 at 37°C (Figures 3A-C). Large scale cell culture (150 ml) was then purified using Ni-NTA column and a single protein band of ~23 kDa was obtained after elution (Figure 3D). The concentration of the purified vif protein was ~173. 3 μg/ml using Bradford assay.
Detection of green fluorescent protein (GFP) expression by fluorescence microscopy and flow cytometry in HEK-293 cells: The DNA constructs (pEGFP-N1,pEGFP-vif) were delivered in vitro by TurboFect. Fluorescence microscopy and flow cytometry were applied to assess GFP expression 48 hr post transfection. According to the obtained data, protein expression was 50% for pEGFP-vif (Figure 4).
Discussion :
Vaccination can play a key role in controlling infectious diseases such as AIDS. To generate an effective therapeutic HIV vaccine, most efforts must be focused on activating cellular immunity 24-26. Virus-specific T cell response has an important role in HIV infection resistance or clearance. Therefore, inducing an immune response against HIV proteins, such as vif protein, may lead to an effective approach 14.
vif has been investigated in some studies to identify effective factors in its expression. In the study by Doi et al, expression level of vif was found to be dependent on nucleotide variations in the vif-coding and also regulatory SA1D2prox region of the genome. They examined clones containing vif-coding sequence of subtype C which expressed low levels of vif/Vif and high levels of vpr/Vpr. They concluded that natural variations of SA1D2prox and vif-coding region might affect the vif expression level and change the replication ability of HIV-1.
In a study by Wang et al, vif expression was found to be at the lowest level in both presence and absence of APOBEC3G (Apolipoprotein B Messenger RNA Editing Enzyme Catalytic Polypeptide 3G) in comparison to other accessory gene evaluations. Moreover, presence of truncated form of vif was identified which was not observed for any other accessory proteins 27.
In the current study, the purpose was designing a recombinant vif protein which is optimized in microbial host and also its expression in mammalian HEK293 cell line in vitro. The gene was cloned and expressed using both pET23a and pEGFP-N1 vectors in BL21(DE3) strain of E. coli and HEK293T cell, respectively for the first time and acceptable expressions were obtained.
vif was expressed by E. coli in an insoluble form probably within inclusion bodies. Sufficient soluble vif after sonication was available for Ni-NTA purification and activity measurements. Despite optimal activity and stability, purified vif contained a small amount of low molecular weight proteins possibly as the result of proteolytic decomposition.
BL21(DE3) expression systems were selected because these cells can be readily manipulated, cultured inexpensively and they grow rapidly. The pET expression system featuring the T7 promoter is by far the most widely used system for heterogeneous expression in E. coli 28. T7 promoter activity is strong and a recombinant protein can accumulate up to 50% of total cellular proteins 29. However, in a study by Khatami et al, origami host of K-12 strain of E. coli was more successful than the BL21(DE3) strain for expressing eukaryotic genes 30.
The optimal expression condition in this study was achieved at OD600 nm of 0.4 with 2.5 mM of IPTG and after an overnight IPTG induction. A final concentration of about 173.7 μg/ml was also obtained by Ni-NTA affinity chromatography.
Kang et al used affinity chromatography and guanidinium chloride, and 5×108 cells were obtained on day 4 after infection 31. Moreover, Andrew and Strebel reached a final concentration of 0.5-1 mg/ml of vif protein expression 32.
For cellular studies, GFP was successfully used as a molecular marker to label vif and thus allowed further studies on its delivery. Therefore, the presence of the vif-GFP fusion protein 24 hr after transfection in the cells (~45%) indicated that the GFP fusion did affect the delivery of vif with TurboFect as a commercial protein transfection reagent in serum-free medium in HEK293 eukaryotic cells. Muller-Taubenberger and Anderson showed that visibility of GFP is highly distinguishable from other fusion tags 33. GFP-positive cells under a fluorescence microscope showed that the linkage of vif to GFP without linker (vif-GFP) was efficient using TurboFect in the mammalian cells.
Conclusion :
The obtained data introduces effective approaches for future therapeutic purposes in vivo and can be used in subsequent HIV vaccine researches.
Acknowledgement :
This study was supported by Department of Hepatitis and AIDS, Pasteur Institute of Iran (Grant no: 983) and was part of an MSc thesis. Our thanks go to Ms. Mona Sadat Laarijani for technical advice.
Conflict of Interest :
The authors who have taken part in this study declare that they do not have anything to disclose regarding the conflict of interest with respect to this manuscript.
Funding: This research was financially supported by Pasteur Institute of Iran (Grant no: 983).
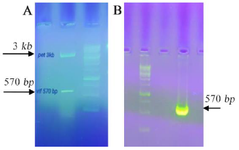
Figure 1. A) Restriction analysis of the plasmid construct in 1% agarose gel. The pET 23a harboring vif was digested by Nhel/Hindlll enzymes. The size of remaining parts of plasmid after digestion was 3608 bp and the size of inserts was 570 bp. B) Confirmation of vif gene cloned in pEGFP-N1/vif vector using colony PCR. The arrow indicates the specific amplified band (570 bp) corresponding the target gene inserted into the vector. DNA size marker of 1 kb was used.
|

Figure 2. Confirmation of the recombinant protein expression by western blotting. Lane 1: un-induced bacterial lysate. Lane 2: bacterial lysate after induction. The pre-stained protein ladder (10-180 kDa) was used.
|
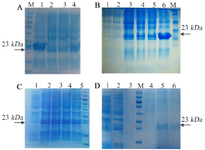
Figure 3. SDS-PAGE analysis for vif protein expression in E. coli BL21 (DE3): (A) Optimum expression to find the best OD600nm for induction; lane 1,2,3 and 4 show 0.4, 0.6, 0.8, and 1, respectively. (B) Time optimization; lane 1: before induction, and lanes 2-6 show 1, 2, 3, 4 hr and O/N, respectively after induction. (C) Optimization of IPTG concentration; lane 1: before induction, and lanes 2-5 show 2.5, 5, 10, and 15 mM, respectively. (D) Recombinant protein purification; lane 1: before induction, lane 2: bacterial crude lysate after induction, lanes 3 - 4: washing solution, lanes 5-6: elution solution. M: protein weight marker is 19-117 kDa. The arrows represent specific 23-kDa band.
|

Figure 4. The GFP expression by immunofluorescence microscope (A and B) and flow cytometry (C). In contrast to un-transfected HEK 293 T cells (B1-B2), the cells co-transfected with pEGFP-N1/vif plasmid produced GFP emission (A1-A2) after 48 hr post-transfection. (C) 41.33% level of expression in GFP in comparison with negative control cells (HEK-293T).
|
|