The Effect of Silver Nanoparticles on Pyocyanin Production of Pseudomonas aeruginosa Isolated From Clinical Specimens
-
Najafi, Mahboobeh
-
Department of Biology, Faculty of Science, Damghan Branch, Islamic Azad University, Damghan, Iran
-
 Yousefi, Ehsan
Department of Biology, Faculty of Science, Mashhad Branch, Islamic Azad University, Mashhad, Iran, Tel: +98 938 1564884, E-mail: e.yousefi@sci.ui.ac.ir
Yousefi, Ehsan
Department of Biology, Faculty of Science, Mashhad Branch, Islamic Azad University, Mashhad, Iran, Tel: +98 938 1564884, E-mail: e.yousefi@sci.ui.ac.ir
Abstract: Background: Pseudomonas aeruginosa (P. aeruginosa) is an opportunistic pathogen causing a wide range of human infections. The organism is resistant to a wide range of antibiotics. The purpose of this study was to investigate the effect of AgNPs on pyocyanin pigment production of P. aeruginosa bacteria isolated from clinical specimens.
Methods: In this study, 15 clinical isolates of P. aeruginosa were collected from different specimens of hospitalized patients. P. aeruginosa was detected by biochemical and molecular (detection of pbo1 gene by colony PCR method) methods and the MIC and MBC of AgNPs were determined by agar dilution method. Inhibition of P. aeruginosa pyocyanin production at AgNPs concentrations of 0, 0.3, 0.5, 1 and 1.5 mg/ml of was studied with OD of 520 nm.
Results: The mean MIC and MBC of AgNPs were 1.229 and 1.687 mg/ml, respectively. Pyocyanin production was investigated for all isolates at different concentrations of nanoparticles, and their comparison showed that with increasing nanoparticle concentration, pyocyanin production significantly decreased (p<0.05).
Conclusion: According to the results of this study, AgNPs had an inhibitory effect on P. aeruginosa and its pigment production and with increasing nanoparticles concentration, pigment production decreased; therefore, it seems that the nanoparticles can be used to treat and prevent diseases caused by P. aeruginosa.
Introduction :
One of the chief global problems is the infections caused by antibiotic-resistant bacteria, and controlling the spread of these infections, especially in health centers, has become a major challenge 1. Pseudomonas aeruginosa (P. aeruginosa) is an opportunistic pathogen causing a wide range of human infections. It is a common hospital-acquired pathogen and responsible for Urinary Tract Infections (UTIs), respiratory infections, dermatitis, soft tissue infections, bacteremia, bone and joint infections, gastrointestinal infections, and a variety of systemic infections, particularly in patients with severe burns, bed ulcers, and in patients suffering from cancer or AIDS who are immunosuppressed 2. The organism is resistant to a wide range of antibiotics 3; at present, the rate of nosocomial infections caused by antibiotic-resistant strains of P. aeruginosa shows a growing trend and is one of the leading causes of death in burn patients 4. In 1995, eighty-eight
thousand people died of nosocomial infections worldwide. Also, P. aeruginosa alone causes 10% of nosocomial infections. Accordingly, in recent years, the causes of nosocomial infections have been considered by many researchers 5. The ability of this bacterium to produce many virulence factors such as phenazines, alginate, proteases, phospholipase C, rhamnolipid, pili (for binding and colonization to host cells) and biofilm formation has made it one of the most important pathogens 6. Pigments produced by P. aeruginosa include pyocyanin (Blue), pyoverdine (Green), pyorubin (Red), and pyomelanin (Black) 7. Pyocyanin (N-methyl, L-hydroxyphenazine) is chemically in the group of phenazines. Phenazines are nitrogen-containing heterocyclic compounds produced by several bacterial species and have been studied for their role in pathogenicity 8. Pyocyanin is involved in biofilm formation as virulence factor; also, in eukaryotic cells as cellular signals, it regulates gene expression that alters cellular responses. Concentrations of pyocyanin in the lungs of patients with chronic cystic fibrosis infection may impair epithelial cell function and reduce the immune response 9. P. aeruginosa has two specific versions of the pyocyanin biosynthesis operon (pbo) called phz A1 B1 C1 D1 E1 F1 G1 (phzI) and phz A2 B2 C2 D2 E2 F2 G2 (phzII). These two operons are 98.3% homologous at the DNA level but different in the promoter region. In addition, two other genes, called phz M and phz S, play a key role in the biosynthesis pathway of pyocyanin 10. With the advent of antibiotic-resistant bacterial strains, researchers today are looking for a new method to treat and control diseases. The science of nanotechnology with a wide range of applications has been able to help researchers in biology and medicine. Silver nanoparticles (AgNPs) with significant antimicrobial effects are one of the most important products of nanotechnology 11,12. These particles by releasing sulfur-containing proteins on the surface of bacterial membranes change their morphology and by changing the bacterial respiratory chain, eventually lead to their death 13. Since bacteria do not become resistant to nanoparticles, they affect a wide range of bacteria 14. In previous research, the antimicrobial effects of the nanoparticles have been proven 15,16. But, research on the effects of nanoparticles on bacterial pigment production is scarce and so far few studies have been conducted on the effect of nanoparticles on the production of P. aeruginosa; moreover, since treatment of bacterial infections has inflicted a lot of cost and problems on the treatment department 17 and given the special importance of pyocyanin in the pathogenesis of P. aeruginosa, the aim of this study was to investigate the antimicrobial effects of AgNPs and their effect on the production of pyocyanin pigment of P. aeruginosa isolated from clinical specimens (with pbo1 gene tracing). In this study, AgNPs synthesized by the US Research Nanomaterials, Inc. were used against the production of pyocyanin. Findings from the present study provide important insights into the potential of AgNPs as an effective new drug for controlling P. aeruginosa-related infections.
Materials and Methods :
Isolation and biochemical identification of P. aeruginosa from clinical specimens: In this study, different specimens of hospitalized patients who were suspected of Pseudomonas infection at Imam Reza and Ghaem Hospitals in Mashhad were transferred to the microbiology laboratory of Islamic Azad University of Mashhad under sterile conditions and next to an ice pack. To isolate P. aeruginosa, cetrimide agar medium and biochemical tests including Gram staining, oxidase, catalase, methyl-red, urea and culture in TSI agar and SIM were used 18.
Molecular identification of P. aeruginosa: Colony Polymerase Chain Reaction (PCR) and pbo1 gene primers (Sinaclon, Iran) were used to confirm P. aeruginosa bacteria isolated from clinical specimens (Table 1). Exact sequences of pbo1 gene were extracted using bioinformatics tools and data from the NCBI database. To ensure the accuracy of the extracted sequences, their evaluation was performed by Blast toolkit at the NCBI database and primers specific for this gene were designed using oligo 7 software. The Blast toolkit was used to ensure that the primers were specific 19.
The colony PCR reaction mixture (Sinaclon, Iran) was prepared with a final volume of 25 µl comprising 3 μl of 1× PCR buffer, 0.75 μl of 0.2 mM MgCl2, 0.5 μl of dNTP Mix (0.5 mM each), 1 μl of 2.5 pmol/μl of each primer, a template DNA, 0.25 μl of Taq DNA polymerase (1.2 U/μl), and 17.5 μl of distilled water. The amplification was carried out in a thermocycler (Kyratec, Korea) with the following cycling conditions: initial denaturation at 95°C for 5 min, and 30 cycles of denaturation at 95°C for 1 min, annealing at 58°C for 1 min, extension at 72°C for 5 min, and final extension at 72°C for 7 min. P. aeruginosa ATCC 1074 (Prepared by Iran’s Scientific and Industrial Research Organization) was used as a positive control, and distilled water as a negative control. The colony PCR products were confirmed by electrophoresis on a 1.5% agarose gel containing 0.5 μg/ml of ethidium bromide (CinnaGen, Iran) in TBE buffer and photographed with UV waves through gel documentation system (Kimiagene, Iran) 20.
Preparation of AgNPs: In this research, AgNPs powder with 99.99% purity, density of 210 nm and average size of 15 nm was obtained from the US Research Nanomaterials, Inc..
Determination of MIC and MBC of AgNPs for P. aeruginosa isolates by agar dilution method: To prepare the bacterial suspension, P. aeruginosa isolates were inoculated in Luria Bertani (LB) (Biomark, India) medium and incubated for 24 hr in the shaker incubator (37°C) (LaCopanion, Korea). To prepare AgNPs suspension, 100 mg of the powder was mixed in 1000 µg of sterile distilled water. Evaluation of Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) of AgNPs were done on a 24-well polystyrene microplate. For this purpose, different concentrations of AgNPs (0 to 50 mg/ml) were prepared in the Mueller-Hinton Agar (MHA) (48°C) (Merck, Germany). After solidification of the culture medium, the bacterial suspension (OD620=0.01) with a volume of 10 μl was inoculated on its surface. Finally, the microplate was incubated for 24 hr at 37°C. After 24 hr, the lowest concentration at which the bacterium did not grow was determined as the MIC. To determine the MBC, the surface of the wells in which the bacterium did not grow was inoculated into culture media without AgNPs and incubated at 37°C for 24 hr. After incubation, the lowest concentration at which the bacterium did not grow was identified as MBC 21. This experiment was repeated 3 times for all isolates and standard strain (P. aeruginosa ATCC 1074) similarly.
Extraction of P. aeruginosa pyocyanin: First, 250 µl of P. aeruginosa [Optical Density at 620 nm (OD620)=0.01] suspension grown in LB medium was inoculated with 25 ml of Glycerol Alanine (GA) (Merck, Germany) medium and incubated for 24 hr in a shaker incubator at 200 rpm. Next, the samples were centrifuged (Sigma, USA) at 4°C and 10,000 rpm for 10 min. The supernatant was collected and passed through a filter with a pore size of 0.22 μm. Next, 4.5 ml of chloroform was added to 7.5 μl of the filtered supernatant and vortexed ten times, each time for 2 seconds. This step was repeated three times for each isolate.
Chloroform was placed at the bottom of the tube; because pyocyanin dissolves in chloroform, it changes to a greenish- blue color. After changing the color, the samples were centrifuged at 10,000 rpm for 10 min. Then, 3 ml of the blue layer collected at the bottom of the tube (Mixture of chloroform and pyocyanin) was transferred to a new tube and in order to acidify the mixture, 1.5 ml of 0.2 N hydrochloric acid was added to it and vortexed (changed blue to pink).
Then, the samples were centrifuged at 10000 rpm for 2 min and 1 ml of pink liquid was transferred to the cuvette and then OD was measured at 520 nm by a spectrophotometer. This experiment was repeated 3 times for all isolates and standard strain (P. aeruginosa ATCC 1074) similarly. The concentration of pyocyanin (μg/ml) was calculated according to the following formula, (OD520 nm×17.072 [molar attenuation coefficient])×1.5 [volume of hydrochloric acid], to determine the dilution factor following transfer into the acidic phase 22,23.
Extraction of P. aeruginosa pyocyanin (Affected by AgNPs): In the first step, 25 ml of GA medium with different concentrations of AgNPs (0 to 1.5 mg/ml) was prepared in erlenmeyer flask and inoculated with 250 μl of P. aeruginosa suspension (OD620=0.01). Erlenmeyer flasks were incubated at 200 rpm for 24 hr (37°C). After the end of the incubation period, the steps of extracting the pyocyanin similar to the previous experiment were performed. This experiment was repeated 3 times for all isolates and standard strain (P. aeruginosa ATCC 1074) similarly.
Statistical analysis: Statistical analysis was performed using SPSS software version 21. Mean±standard deviation was obtained by one-way ANOVA and p-values of less than 0.05 (p<0.05) were considered statistically significant.
Results :
Biochemical and molecular identification of P. aeruginosa: Based on the results of biochemical tests in table 2, 15 P. aeruginosa isolates were collected from clinical specimens. PCR results of pbo1 gene confirmed all 15 P. aeruginosa isolates by biochemical tests (100% of clinical specimens). In figure 1, a gel electrophoresis PCR product of several isolates is shown next to the 100-bp marker.
Determination of MIC and MBC of AgNPs: The mean (±standard deviation) MIC and MBC of AgNPs for P. aeruginosa isolated from clinical specimens were 1.229±288 and 1.687±288 mg/ml and for P. aeruginosa strain ATCC 1074 were 1.333±288 and 1.333±288 mg/ml, respectively (Table 3).
There was no significant difference in MIC of P. aeruginosa strains isolated from clinical specimens and standard strains (p<0.05).
Extraction of P. aeruginosa pyocyanin: The average OD of P. aeruginosa pyocyanin after extraction without the influence of AgNPs and at different concentrations of AgNPs is shown in table 4.
All isolates could produce pyocyanin; mean OD of P. aeruginosa pyocyanin at 520 nm after extraction (without the effect of AgNPs) was 0.584 and the mean OD of P. aeruginosa pyocyanin at 520 nm after extraction with concentrations of 0 (Control), 0.3, 0.5, 1, and 1.5 mg/ml was 0.556, 0.277, 0.109, 0.09 and 0, respectively. The highest OD (OD520) at 0.576 nm was related to isolate 10 at a concentration of 0.3 mg/ml and all isolates at a concentration of 1.5 mg/ml had an OD equal or close to zero.
Discussion :
The effect of AgNPs on pyocyanin production was investigated in 15 P. aeruginosa isolates of clinical specimens. Based on results of ANOVA, it was found that with increasing the concentration of AgNPs, the amount of OD of pyocyanin produced by P. aeruginosa and standard strains was significantly reduced compared to the control (Concentration 0) (p<0.05) which shows the inhibitory effect of AgNPs on the production of P. aeruginosa pyocyanin. The results of OD at 520 nm showed that the ability to produce pyocyanin in the clinical strains isolated in this study is higher than the standard strain. Finnan et al 24 isolated 12 P. aeruginosa bacteria from patients with cystic fibrosis and determined the frequency of phzM, phzII, phzH and phzS genes to be 91.6%, 100%, 100% and 8.3%, respectively; phzII and phzH genes like pbo1 gene in the present study were detected in 100% of the clinical specimens. In another study by Dadmanesh et al 25 in 2013, the frequency of phzI, phzII, phzS, and phzM genes in 23 P. aeruginosa bacteria isolated from the urine of patients with urinary tract infections was 11.7, 28.4, 19.6, and 36.2 %, respectively. The frequency of all genes in their study was lower than the frequency of pbo1 gene in our study. This difference may be due to research on a specific clinical specimen (Urine only). In 2016, Nasiri et al 26 reported 1 mg/ml MIC of AgNPs against P. aeruginosa isolated from urine cultures of hospitalized patients. Their MIC values were slightly lower than the ones in the present study. This difference in the two studies may be due to differences in the type of specimens tested. In a study, Khan et al 27 showed that the production of P. aeruginosa PAO1 KCTC 1637 pyocyanin at gold nanoparticles concentrations of 0.032, 0.128 and 0.256 mg/ml decreased 79.4, 81.9, and 87.7%, respectively compared to the control. The results of their study, similar to the results of the present study, proved the effects of nanoparticles on reducing the production of P. aeruginosa pyocyanin. P. aeruginosa virulence is a multifactorial process and has been attributed to cell associated factors or secreted virulence factors, pyocyanin and phenazine operons (phzI and phzII); the genes encode precursor proteins involved in the formation of three phenazine compounds (phzH, phzM and phzS) 25. Therefore, reduction of pyocyanin can be considered to effectively attenuate the pathogenesis and colonization of P. aeruginosa without affecting bacterial growth or initiating resistance selection. Blue-green pigment pyocyanin essentially causes oxidative stress and cytotoxicity to the host tissues 27.
Conclusion :
According to the results of this study, AgNPs had an inhibitory effect on P. aeruginosa and its pigment production and with increasing nanoparticle concentration, pigment production decreased. AgNPs may be a good alternative to antibiotics and chemical drugs and can be used to treat and prevent diseases caused by P. aeruginosa. Therefore, it is necessary to extend the scope of research to investigate the effects of nanoparticles on the production of P. aeruginosa pigments in the prevention and treatment of diseases caused by them and to prevent the emergence of antibiotic-resistant strains. Further studies should assess the combination of AgNPs and antibiotics against resistant strains for the development of new materials and substances for medical application.
Acknowledgement :
We would like to thank the microbiology laboratory of Ghaem and Imam Reza Hospitals and the Islamic Azad University of Mashhad and all those who helped us with this research. We had no sources of financial support.
Conflict of Interest :
There is no conflict of interest.
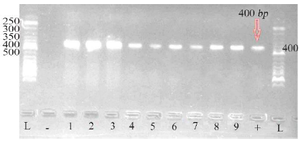
Figure 1. Gel electrophoresis of PCR product of pbo1 gene from several P. aeruginosa isolates along with the standard 50-bp marker (1-9: Samples; L: Ladder; +: Positive control; -: Negative control).
|

Table 1. Characteristics of primers used to detect pbo1 gene in P. aeruginosa isolates
|

Table 2. Results of biochemical tests to detect P. aeruginosa
|
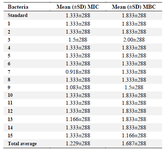
Table 3. Mean MIC and MBC of AgNPs for 15 clinical isolates of P. aeruginosa and standard strains (data in mg/ml) (Repeated three times)
|
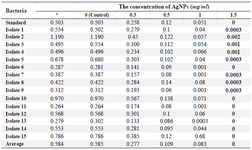
Table 4. OD of pyocyanin produced by clinical isolates of P. aeruginosa and the effect of different concentrations of AgNPs on the production of pyocyanin in these isolates based on OD at 520 nm
*: OD of pyocyanin produced without the effect of AgNPs.
|
|