Role of Phosphorylation and Hyperphosphorylation of Tau in Its Interaction with βα Dimeric Tubulin Studied from a Bioinformatics Perspective
-
Dixit , Hrushikesh
-
Faculty of Biotechnology and Bioinformatics, D.Y.Patil Deemed to be University, CBD Belapur, Navi Mumbai, India
-
 Kumar C, Selvaa
Faculty of Biotechnology and Bioinformatics, D.Y. Patil Deemed to be University, CBD Belapur, Navi Mumbai, India, Email: selvaakumar.c@dypatil.edu
Kumar C, Selvaa
Faculty of Biotechnology and Bioinformatics, D.Y. Patil Deemed to be University, CBD Belapur, Navi Mumbai, India, Email: selvaakumar.c@dypatil.edu
-
Chaudhary, Ruchi
-
Faculty of Biotechnology and Bioinformatics, D.Y.Patil Deemed to be University, CBD Belapur, Navi Mumbai, India
-
Thaker, Divya
-
Faculty of Biotechnology and Bioinformatics, D.Y.Patil Deemed to be University, CBD Belapur, Navi Mumbai, India
-
 Gadewal, Nikhil
Advanced Centre for Treatment, Research and Education in Cancer (ACTREC), Kharghar, Navi Mumbai, India, Email: ngadewal@actrec.gov.in
Gadewal, Nikhil
Advanced Centre for Treatment, Research and Education in Cancer (ACTREC), Kharghar, Navi Mumbai, India, Email: ngadewal@actrec.gov.in
-
 Dasgupta, Debjani
Faculty of Biotechnology and Bioinformatics, D.Y. Patil Deemed to be University, CBD Belapur, Navi Mumbai, India, Email: drdebjanid@dypatil.edu
Dasgupta, Debjani
Faculty of Biotechnology and Bioinformatics, D.Y. Patil Deemed to be University, CBD Belapur, Navi Mumbai, India, Email: drdebjanid@dypatil.edu
Abstract: Background: Tau is a disordered Microtubule Associated Protein (MAP) which prefers to bind and stabilize microtubules. Phosphorylation of tau in particular enhances tau-tubulin interaction which otherwise detaches from tubulin during hyperphosphorylation. The reason behind their destabilization, detachment and the role of β subunit (from microtubule) and the projection domain (Tau) in microtubule stability remains elusive till date. Thus, a complete 3D structural investigation of tau protein is much needed to address these queries as the existing crystal structures are in fragments and quite limited.
Methods: In this study, the modelled human tau protein was subjected to phosphorylation and hyperphosphorylation which were later considered for docking with microtubules (βα subunits-inter dimer) and vinblastine.
Results: Phosphorylated tau protein interacts with both α- and β subunits. But stronger bonding was with α- compared to β subunits. Regarding β subunit, proline rich loop and projection domain actively participated in tau binding. Interestingly, hyperphosphorylation of tau increases MAP domain flexibility which ultimately results in tau detachment, the main reason behind tangle formation in Alzheimer’s disease.
Conclusion: This study being the first of its kind emphasizes the role of projection domain and proline rich region of β-subunit in stabilizing the tau-tubulin interaction and also the effect of hyperphosphorylation in protein-protein and protein-drug binding.
Introduction :
Tau, a Microtubule Associated Protein (MAP), is critically involved in assembly, stability, dynamics, and maintenance of axonal structure 1,2. The role of structure and function of tau protein has been implicated in several neuronal diseases especially Alzheimer’s disease 3. Tau being an intrinsically disordered protein interacts and stabilizes microtubules 4,5. Tau has eight distinct domains classified into N-terminal domain (N1, N2), proline rich domain (P1 and P2) and four microtubule-binding domains. A combination of N1, N2 and P1 domains makes up the projection domain whereas the combination of repeat domain 1,2,3,4 (R1:561-591; R2:592-622; R3:623-653; R4:654-685) comprises the MT binding domain 6 (Figure 1). It is known that, repeat domain and proline rich domain strongly interacts with microtubule 7. The role of projection domains and their affinity towards microtubule have remained controversial till date 8. The negatively charged MAP domain of microtubule prefers to interact with the positively charged C-terminal Microtubule Binding Domain (MBD) (Residues 561-685) 9. This MBD is made of three or four binding domains of which, the four binding domains demonstrated better microtubule stability compared to three domains 10. There is weak interaction between assembly domain and microtubule which is strengthened by the proline rich domain 11,12. Alternative splicing in tau gene (MAPT) produced six isoforms which differ from others in the presence of the N-terminal inserts, N1 and N2, and the second repeat domain, R2. These three domains are encoded by exon 2, 3 and 10 13.
Microtubules, the preferred partner of tau, are involved in cell division, chromosomal segregation, motility and intracellular transport 14,15. These polymers are made up of heterodimeric α and β subunits arranged in a lateral and longitudinal fashion. The arrangement of α and β subunits in a head-to-tail fashion forms inter-dimer (β-α) and intra-dimer (α-β) protofilament 16-20. Tau prefers to interact with inter-dimeric region (β-α) which is also the binding pocket for vinblastine (Figure 2). Additionally, tau also reaches the interior of the microtubule near the paclitaxel binding site 21-24. Post translational modifications like glycation, glycosylation, ubiquitination, phosphorylation, truncation and nitration in tau protein have been well investigated 25. Phosphorylation, in particular, helps in gaining protein secondary structures and also assists in tau-microtubule binding. After hyperphosphorylation, tau detaches from tubulin and gets tangled during self-assembly leading to Alzheimer’s disease 26-28. Tau being a highly disordered protein, is available in fragments in Protein Data Bank instead of complete structure. The interaction between the non-phosphorylated tau with tubulin protein was already explored from in-silico perspective 29. To date, the role of phosphorylation and hyperphosphorylation of tau protein and their effect on microtubule and drug binding remains unclear. In this novel study, the effect of phosphorylation and hyperphosphorylation of modelled tau protein on microtubule and vinblastine binding was investigated for the first time.
Materials and Methods :
3D modeling human tau protein with 758 amino acids was downloaded from Uniprot Database 30 (ID: P10636) and selected as a query sequence to identify potential template using BLAST-PDB online server 31. I-TASSER (Iterative Threading ASSEmbly Refinement) online server 32 based on ab inito and threading method was considered for complete model building. Thus, the top 10 threading templates were selected using LOMETS by means of multiple threading approaches. Furthermore, generated decoys were clustered based on the pair-wise structure similarity using SPICER program. As a result, the top five largest structure clusters listed their respective five models (Model 1-5) which were considered for confidence score measurement through C-score and TM score. To begin with, C-score relies on the significance of threading template alignments wherein the highest value implies the highest confidence score. Next, TM score was used to measure the structural similarity between two structures to address the problem of Root Mean Square Deviation (RMSD) which is quite sensitive to local errors. Most importantly, the highest TM score confirms correct topology for the 3D model. From the top five models, the modelled tau structure with best Tm-align (More than 0.5) and highest C-score was selected for Phosphorylation (PT) and Hyperphosphorylation (HPT) using Vienna-PTM 2.0 webserver (http://vienna-ptm.univie.ac.at) 33. Phosphor group was added to the selected serine and threonine residues of the modelled tau protein. As per the literature review, six positions (Serine: 516, 519, 579,739) and (Threonine: 522, 548), were considered 34-40 for phosphorylation, of which 516, 519, 522 and 548 are from proline rich domain, 579 from Repeat 1, and 739 from C-terminal region. For HPT, fifty sites were identified as listed by the software 41 (Supplementary information table 1). Next, αβ tubulin dimer was downloaded from Protein Data Bank 42 (PDB ID: 4O4H) 43 which was needed for docking with modelled tau. The downloaded crystal structure with a resolution of 2.1Å had four chains (A, B, C, and D) intact of which only B: β chain C: α chain of inter-dimer (β-α) were separated using Swiss Pdb-Viewer 44 as they were the regions of tau binding based on the literature review 45. β-α tubulin subunits were subjected to energy minimization using GROMACS 4.5.5 46 standalone software. The cubic box size was 1.0 nm from the box edge. In total, the minimization steps were 50000 and the minimization step size was 0.01.
Simulation of phosphorylated tau (PT) and hyperphosphorylated tau protein (HPT): Modelling of tau protein was quite a challenging task due to its disordered nature and limited structural details available in Protein Data Bank. Modeled tau has shown stereochemically unfavourable residues in the 3D structure. To optimize this, the modelled PT and HPT proteins were subjected to molecular dynamics simulation using GROMACS 4.5.5. All the simulations were run on Tesla K80 GPU based on Linux workstation. The systems were solvated using TIP3P water model 47 in a cubic box with periodic boundary conditions. The cubic box size of water surrounding the dimers was 1.2 nm. Considering 1.2 nm buffer between the outside of the dimer and the edge of the box, the total buffer space was 2.4 nm between the cells of the periodic boundary conditions. Taking into account the short-range van der Waals forces and electrostatic cut-offs of 0.8 nm, the extra space of 0.4 nm would be sufficient for the dimers if they unfolded during simulation. Furthermore, monovalent counterions like Na and Cl ions were added to neutralize the system. The systems were first energy minimized using the steepest descent algorithm with a tolerance of 1000 kJ/mol/nm. Long-range electrostatic interactions were calculated using Particle Mesh Ewald (PME) summation with 1 nm cut-offs for Coulombic interactions, and van der Waals interactions were calculated with a distance cut-off of 1.4 nm. Later, the systems were equilibrated by applying positional restraints on the structure using NVT followed by NPT ensemble for 100 ps each. The temperature of 300 K was coupled using a Berendsen thermostat 48 with pressure at 1 bar, coupled by the Parrinello-Rahman algorithm 49. The equilibrated systems were then subjected to 100 ns of production run with time-step integration of 2 fs. The trajectories were saved at every 2 ps and analyzed using analysis tools from GROMACS 4.5.5. The Root Mean Square Deviation (RMSD), Root Mean Square Fluctuation (RMSF) and Radius of Gyration (Rg) were analyzed individually for the phosphorylated and hyperphosphorylated tau protein in association with βα subunits. RMSD helps in calculating the average change in the displacement of a selection of atoms for a particular frame with respect to a reference frame. It also indicates whether the simulation has equilibrated or not. RMSF is mainly used for illustrating local changes in the protein chain through peaks that fluctuate the most during the simulation 50. Regarding Rg, it is an indicator of protein structure compactness 51. After simulation, the stereochemically stable PT and HPT proteins with least potential energy were selected and validated using PROCHECK-Ramachandran plot and ProSA-web server. The secondary structure of the phosphorylated tau protein was compared with the available crystal structures from Protein Data Bank.
Docking of β-α tubulin with PT and HPT proteins: Both PT and HPT proteins were docked with inter-dimer β-α subunits using ClusPro online server 52. Three steps were followed for complex generation which includes rigid body docking, Root Mean Square Deviation (RMSD) based clustering and energy minimization based refinement of selected structures. β-α subunits and tau protein were selected as receptor and ligand, respectively using DOCK option. According to literature review, proline rich region along with microtubule binding site of tau protein interacts with the C-terminal MAP domain (381-440) of the tubulin protein. Thus, poses were selected strictly based on these criteria. ClusPro docked complexes were purely based on cluster population rather than their energy value. As a result, the binding energy of the best pose of PT-βα and HPT-βα was calculated using dDFIRE server 53.
Simulation of docked complex of PT and HPT protein with tubulin inter-dimer: The docked complexes of PT-β-α subunits and HPT-β-α subunits were subjected to molecular dynamics simulation using GROMACS 4.5.5. All these simulations were run on Tesla K80 GPU based on Linux workstation with similar steps mentioned above. After simulation, the RMSD, RMSF and Rg plots were generated and analyzed for the overall domain rigidity and flexibility.
Docking of vinblastine with PT-βα and HPT- βα complex: Based on literature review, tau competes with vinblastine during microtubule interaction which needed further introspection from in-silico perspective 21-23. For the same reason, β-α subunits, PT-β-α subunits and HPT-β-α subunits were considered for docking with vinblastine obtained from Protein Data Bank (PDB id: 4EB6) 54 using Glide tool of Schrodinger suite 55. LigPrep tool of Schrodinger was used to generate 28 conformers of vinblastine based on protonation states at pH=7.0+/-2.0 using Epik. The docking site on the tubulin protein within the complexes was generated using receptor-grid generation protocol of Glide by selecting the residues which are α tubulin: 240-252;327-341; 348-353 and β tubulin: 212-215; 172-181; 215-223 22. A scaling factor of 1.0Å was set to van der Waals (VDW) radii for the atoms of residues that probably interact with ligands with the partial atomic charge cut-off of 0.25Å. The scaling factor of van der Waals radii for ligand was set to 0.80 Å and partial charge cut-off of 0.15A. An extra precision (XP) mode of Glide docking was done which does extensive sampling and provides reasonable binding poses. Post-docking, the minimization of docked complexes, was carried out to obtain the glide score.
Results :
Based on ab initio and threading method, human tau protein was modelled wherein ten templates were selected and considered for simulation which ultimately resulted in 600 decoys. They were later clustered using SPICER program based on the pair-wise structure similarity. Thus, five largest structure clusters listed five best models of which, model1 with a C-score and TM score of 0.55 and 0.79±0.09, respectively was selected for phosphorylation and hyperphosphorylation. The rest four models with a negative C-score without any TM score were ignored.
Simulation of PT and HPT proteins: As per the Cα RMSD plot analysis, both phosphorylated and hyperphosphorylated tau are similar till 40 ns. After 40 ns, hyperphosphorylation in tau has led to an increase in the RMSD by approximately 1 nm as compared to phosphorylated tau, which clearly confirms that there is a considerable structural change within the hyperphosphorylated tau protein (Figure 3A). Comparative RMSF plot analysis of the projection domain, proline rich domain and the repeat domain showed higher flexibility in the hyperphosphorylated tau protein. Phosphorylated tau protein was moderately flexible in nature except for Repeat 3 (Table 1) (Figure 3B). The Rg of the proteins also corroborated with the RMSD, where distinct increase in Rg of hyperphosphorylated tau is due to less compact structure as compared to phosphorylated tau protein (Supplementary information, figure S1). Phosphorylated and hyperphosphorylated tau protein with the least potential energy of -3283161.50 kJ/mol and -3282928.00 kJ/mol, respectively were selected and considered for structure validation (Figure 4A). As per PROCHECK-Ramachandran plot report, 87.1% of residues were in the most favored region; 10.5% were in addition allowed region, 1.4% in generously allowed region and 1% in disallowed region (Figure 4B). Regarding ProSA-Web report, the overall model quality was within the permissible limit. As far as the local model quality was concerned, the graph with a window size of 40 amino acids was on the negative side confirming their structural stability (Supplementary information, figure S2). The secondary structures of the modelled tau with the least potential energy was compared with the available crystal structures. The PDB ID: 5N5A 56 and 4TQE 57 showed better secondary structural similarity with the modelled tau protein (Supplementary information, figure S3).
Protein-protein docking: ClusPro generated docked poses for PT-βα and HPT-βα complexes were categorized into balanced, electrostatic-favored, hydrophobic-favored and van der Waals and electrostatic states. Of these, thirty balanced poses were downloaded and individually visualized using CHIMERA 1.131 58 to identify their region of interaction as per literature review. The best poses for PT-βα and HPT-βα in association with MAP domain of tubulin and the assembly domain of tau proteins were selected for binding affinity using dDFIRE software. As per the software report, the binding energies of PT with β and α subunits were -1867.58 kcal/mol and -1907.94 kcal/mol, respectively. Conversely, the binding affinity of HPT with β and α subunits was -1657.83 kcal/mol and -1788.20 kcal/mol, respectively. Interestingly, α subunit of tubulin demonstrated better binding affinity with PT and HPT protein compared to the β subunit. Furthermore, the overall difference in binding energy of α subunit with PT and HPT was -119.74 kcal/mol whereas the difference in binding affinity of β subunit with PT and HPT was -209.75 kcal/mol. Thus, it was quite evident that after hyperphosphorylation, the interaction between tau and α subunit was not much affected as compared to that of β subunit with tau.
The hydrogen bonds between the PT-βα and HPT-βα subunits were investigated from structural perspective. In PT-βα complex, mostly MAP domain of α subunit interacts with the Repeat 2 region of tau through salt bridge formations. Only a single hydrogen bond formation was observed between the projection domain of PT protein and the MAP domain of α subunit. In particular, hydrogen bonds were observed between H10; H10-B9 and the proximal region of the projection domain which are the integral part of the longitudinal tubulin interaction 23. β subunit interacts with proline rich region (Arg448) and the projection domain (Glu127, Lys130, Glu134, Asp177). Thus, β subunit has no interaction with repeat domain in phosphorylated tau-tubulin interaction. Most importantly, weak binding was observed between the projection domain, proline rich region and the β subunit compared to the repeat domain and α subunit. As per our findings, acidic amino acids of α subunit (Glu433, Glu429, and Asp392) interact with basic amino acid (Lys607 and Lys611) of phosphorylated tau protein which is in agreement with the previous literature 9. The other saltbridge was between the Lys311 (S8) of α subunit and Glu161 (Projection domain) of PT. Thus, α subunit interacts with Repeat 2 (Lys611, Lys607, Asn612 and Asn613) and the projection domain (Arg138, Glu161 and His190) of the tau protein (Table 2). Thus, four salt bridges were observed between the β subunit and the PT protein and five salt bridges between α subunit and PT protein. This unique finding during our study justifies that the binding affinity of PT protein for α subunit is much stronger than for the β subunit. In total, nine salt bridges were observed between the phosphorylated PT and βα subunits (Figures 5A and B). However, after hyperphosphorylation, the α alpha subunit lost its contact with the Repeat 2 of tau which ultimately resulted in lower binding affinity. A single salt bridge remained between Lys176 of β subunit with Glu186 in HPT protein (Figures 5C and D). To summarize, after hyperphosphorylation, the βα subunits lost their contact with projection domain, proline rich domain and Repeat 2 domain of tau protein. The docked protein-protein complexes of PT-βα and HPT-βα are shown in supplementary information, figure S4.
Protein-protein simulation: Both PT and HPT were docked with βα tubulin subunits and considered for simulation. Based on the RMSD report, PT-βα equilibrated at 42 ns with 1.41 nm deviation after phosphorylation. However, after hyperphosphorylation, HPT-βα attained its equilibration at 46 ns with a standard deviation of 1.8 nm (Figure 6A). Thus, hyperphosphorylated tau undergoes considerable structural changes evident from the change in standard deviation which has a direct impact on protein-protein interaction. As per the RMSF plot for tau protein, the projection domain and the proline rich domain becomes flexible after hyperphosphorylation. However, Repeat1 and Repeat 4 are rigid in HPT compared to the PT protein (Figure 6B). As per the RMSF plot of α and β subunit, the helix H10 and H10-S9 domains critically involved in longitudinal interaction are highly flexible in β subunit after hyperphosphorylation (Figure 7A). Moreover, these domains were stable in native and phosphorylated tau protein. Highly flexible native α subunit becomes rigid in PT-βα and HPT-βα complexes (Figure 7B). It was observed that the MAP domain of β-HPT displayed a maximum flexibility, whereas α subunit does not show any substantial structural changes in native α, or in α-PT and α-HPT complexes (Figures 7C and D) (Table 3). Thus, phosphorylation and hyperphosphorylation had lesser impact on the stability of α subunit as compared to β tubulin. Rg plot confirms less compact structure after hyperphosphorylation (Supplementary information, figure S5).
Protein-ligand docking: The stable conformer of vinblastine was docked with native βα, PT-βα and HPT-βα within the inter-dimeric region. As per the docking report, vinblastine showed a binding energy of -6.007 kcal/mol and -7.99 kcal/mol with native βα and PT-βα protein complex, respectively. There were no interactions between drug and HPT-βα complex. Interactions between the drug and tau-tubulin complex were investigated separately. As per our findings, in βα subunits, vinblastine interacts with Asn329 and Val353 of α subunit; Asp179 and Pro222 of β subunit. In particular, Asn329 from helix H10 is indispensable for lateral and longitudinal interaction. Additionally, Val353 from S9 domain is also a preferred site for the drug. Similarly, vinblastine also interacts with Asp179 of Ribose binding loop and Pro222 of H6-H7 loop of β subunit. However, after phosphorylation, an additional hydrophobic interaction was observed between Val352 and the drug in α subunit without any change in their binding pattern with β subunit (Figures 8A and B). Interestingly, after hyperphosphorylation, vinblastine completely lost its contact with tubulin protein.
Discussion :
The soluble and highly disordered tau protein plays a crucial role in the maintenance of axonal structure. As per literature review, post translational modifications like phosphorylation and hyperphosphorylation result in detachment from tubulin protein which ultimately causes the self-aggregation of tau protein and disease. Understanding the overall effect of post translational modification required the 3D structure of tau protein which was modelled and considered for phosphorylation and hyperphosphorylation. As per the literature review, an overall increase in tau phosphorylation reduces its affinity towards microtubules which brings in destabilization of neuronal cytoskeleton 26-28. Based on our in-silico findings, both β and α subunits showed a strong interaction with phosphorylated tau protein leading to their structural stability. This study being the first of its kind from structural perspective agrees with the statement put forth by Hong, Hutton and Lee et al 26-28. Interestingly, our study indicates that on hyperphosphorylation, the interaction of tau with β subunit is reduced far more than its interaction with α tubulin, till it finally detaches from the microtubules. This corroborates with the observations made by Martin et al 59. Repeat regions critically involved in both microtubule interaction and their stabilization as per literature review 60 become rigid after hyperphosphorylation. Furthermore, overall flexibility of the MAP domain and the lateral-longitudinal contacts like helix H10 and H10-B9 loop in β subunit which remain rigid in α subunit could play a crucial role in the detachment of the tau protein. Our findings confirm the role of salt bridges during phosphorylation which strengthens the interaction between tau and βα tubulin. However, after hyperphosphorylation, loss in salt bridges results in their detachment from tubulin.
Conclusion :
Both tau and vinblastine compete with helix H10 of the inter-dimeric region of microtubule which is essential for lateral and longitudinal contact. Importantly, tau stabilizes the interaction between microtubules which get destabilized after their interaction with vinblastine. As per our findings, in presence of hyperphosphorylated tau, vinblastine ceases to interact with the inter-dimer interface. Thus to summarize, it has been shown here for the first time that tau interacts with βα dimer wherein the binding affinity is stronger with α subunit compared to β subunit. Furthermore, tau detaches from the β subunit due to the higher flexibility of the MAP domain and its loss of contact with projection and proline rich domains. This study is the first attempt to understand the dynamics of tau-tubulin interaction which helps in understanding Alzheimer’s disease from a structural perspective.
Acknowledgement :
The authors would like to acknowledge the Department of Biotechnology (DBT), Government of India, sponsored Distributed Information Sub Centre (SubDIC) of Biotechnology Information System (BTIS) Network at ACTREC where the analysis of MD trajectories and docking studies was carried out. Lastly, the authors would like to thank the management of DY Patil Deemed to be University for providing the faculties to do the in-silico studies. This project was not funded by any external funding agencies.
Conflict of Interest :
There are no conflicts of interest.

Figure 1. Schematic diagram of tau protein with its projection domains, proline rich domain, microtubule binding region and C-terminal region.
|
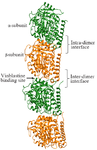
Figure 2. The tubulin dimer with intra- (αβ) and inter-dimer (βα) subunits. The drug vinblastine binding site is highlighted using an arrow.
|
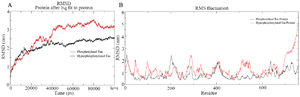
Figure 3. A) Combined Root Mean Square Deviation of the phosphorylated and hyperphosphorylated tau protein. The phosphorylated tau plot showing stable deviation plot is shown in black color. The hyperphosphorylated tau displaying structural changes during simulation is shown in red colour. B) Combined Root Mean Square Fluctuation of the phosphorylated and hyperphosphorylated tau protein. Higher mobility is experienced in projection domain, proline rich domain and the repeat domain of hyperphosphorylated tau protein.
|
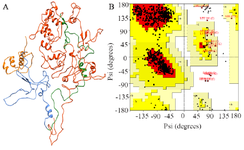
Figure 4. A) Modeled tau protein with its four domains, B) Ramachandran plot of modeled tau protein.
|
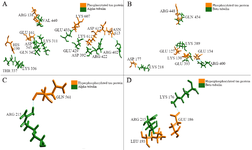
Figure 5. Phosphorylated and hyperphosphorylated tau interaction with βα subunits, A) Four salt bridges observed between the phosphorylated PT and β subunits, B) Five salt bridges observed between the phosphorylated PT and α subunits, C) No salt bridge observed between HPT and β subunits, D) Single salt bridge observed between the phosphorylated PT and α subunits.
|

Figure 6. A) Combined Root Mean Square Deviation of the phosphorylated and hyperphosphorylated tau protein with tubulin dimer. The phosphorylated tau with tubulin showing stable deviation plot is shown in black color. The hyperphosphorylated tau with tubulin displaying structural changes during simulation is shown in red color, B) Combined Root Mean Square Deviation of the phosphorylated and hyperphosphorylated tau protein with tubulin dimer. Overall domain mobility of the projection domains, proline rich domain, microtubule binding region and C-terminal region is displayed through plot.
|

Figure 7. Combined RMSF plot of α and β subunit with tau involved in longitudinal interaction. Highly flexible native α subunit which becomes rigid in PT-βα, (A) HPT-βα, (B) complexes. No structural changes of MAP domain were observed in the native α, or in α-PT, (C) and α-HPT complexes (D). Maximum conformational changes were observed in the MAP domain of β-HPT.
|
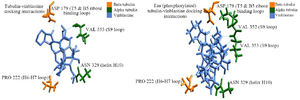
Figure 8. Vinblastine binding with native βα, PT-βα and HPT-βα, A) vinblastine interaction with native βα, B) vinblastine interaction with PT-βα.
|
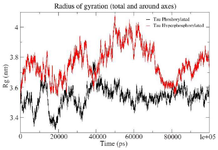
Figure S1. Combined Radius of Gyration of the phosphorylated and hyperphosphorylated tau protein. An increase in Rg is observed in hyperphosphorylated tau protein due to less compact structure.
|
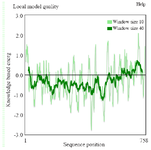
Figure S2. ProSA-Web showing the local model quality of the modeled tau protein which was generated with a window size of 40 amino acids.
|
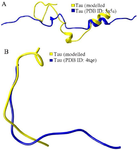
Figure S3. As per the secondary structure comparison, modeled tau protein showed better structural resemblance in S3(A) and S3(B) with PDB ID: 5N5A and 4TQE, respectively.
|
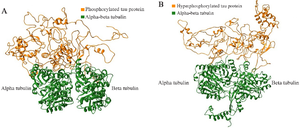
Figure S4. The complete docked protein-protein complexes of PT-βα and HPT-βα are shown in ribbon file format as S4 (A) and S4 (B), respectively.
|
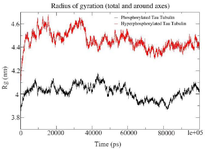
Figure S5. Combined Radius of Gyration of the phosphorylated and hyperphosphorylated tau protein with tubulin. An increase in Rg is observed in hyperphosphorylated tau protein due to less compact structure.
|
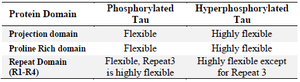
Table 1. Root Mean Square fluctuation in PT and HPT
|
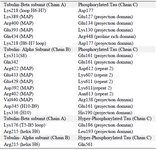
Table 2. Interactions between PT-βα and HPT-βα protein. Amino acids and their respective domain were mentioned for β and α subunits along with tau protein
|

Table 3. Root Mean Square fluctuation in PT-βα and HPT-βα
|
Supporting information table 1. List of residues phosphorylated using Vienna-ptm 2.0
|
|