Silencing SALL-4 Gene by Transfecting Small Interfering RNA with Targeted Aminoglycoside-Carboxyalkyl Polyethylenimine Nano-Polyplexes Reduced Migration of MCF-7 Breast Cancer Cells
-
Noruzi, Somaye
-
Department of Advanced Sciences and Technologies, Faculty of Medicine, North Khorasan University of Medical Sciences, Bojnurd, Iran
-
Student Research Committee, Faculty of Medicine, North Khorasan University of Medical Sciences, Bojnurd, Iran
-
Vatanchian, Mehran
-
Department of Anatomical Sciences, Faculty of Medicine, North Khorasan University of Medical Sciences, Bojnurd, Iran
-
Azimian, Amir
-
Department of Pathobiology and Laboratory Sciences, Faculty of Medicine, North Khorasan University of Medical Sciences, Bojnurd, Iran
-
Niroomand, Arash
-
Department of Advanced Sciences and Technologies, Faculty of Medicine, North Khorasan University of Medical Sciences, Bojnurd, Iran
-
Student Research Committee, Faculty of Medicine, North Khorasan University of Medical Sciences, Bojnurd, Iran
-
 Salarinia, Reza
Department of Advanced Sciences and Technologies, Faculty of Medicine, North Khorasan University of Medical Sciences, Bojnurd, Iran, Tel: +98 58 32297095, E-mail: rezasalarinia@gmail.com
Salarinia, Reza
Department of Advanced Sciences and Technologies, Faculty of Medicine, North Khorasan University of Medical Sciences, Bojnurd, Iran, Tel: +98 58 32297095, E-mail: rezasalarinia@gmail.com
-
 Oroojalian, Fatemeh
Department of Advanced Sciences and Technologies, Faculty of Medicine, North Khorasan University of Medical Sciences, Bojnurd, Iran, Tel: +98 58 32297095, E-mail: f.oroojalian@ut.ac.ir, f.oroojalian@gmail.com
Oroojalian, Fatemeh
Department of Advanced Sciences and Technologies, Faculty of Medicine, North Khorasan University of Medical Sciences, Bojnurd, Iran, Tel: +98 58 32297095, E-mail: f.oroojalian@ut.ac.ir, f.oroojalian@gmail.com
Abstract: Background: The application of non-viral systems for delivering genes to cells is becoming a very interesting issue, especially in the treatment of neoplasms such as Breast Cancer (BC). Polymer-based non-viral systems are safe and feasible gene carriers to be used in targeted cancer therapy. SALL4 gene encodes a transcription factor and is overexpressed in some cancers.
Methods: In this study, carboxyalkylated-PEI25 (25 kDa) was used to deliver plasmids expressing SALL4-siRNA into MCF-7 cells. DLS and AFM were applied to determine the size of nanoparticles. The MTT method was used to assess cytotoxicity, and the efficiency of transfection was confirmed both qualitatively and quantitatively. Finally, the effect of silencing SALL4 was investigated on the migration of MCF7 cells using the scratch test.
Results: The results showed that transferring the SALL4-siRNA using PEI25G10C50 reduced the expression of the corresponding transcription factor by 14 folds which attenuated the migration of MCF-7 cells by 58%.
Conclusion: In conclusion, PEI25G10C50 can serve as an effective gene delivery system for treating BC by targeting SALL-4.
Introduction :
Breast Cancer (BC) is the most frequent type of cancer and cause of cancer related mortality among women worldwide 1. This cancer is commonly treated by either radiotherapy, surgery, systemic hormone therapy, chemotherapy, or combinations of these 2. Nevertheless, the effectiveness of these treatments cannot be warranted, and they may be associated with long-term serious side effects and reduced patients’ quality of life, particularly in the case of metastatic BC 3. On the other hand, the development of immune escape and drug resistance mechanisms, as well as metastatic features in cancer cells causes a high rate of treatment failure in cancers 4. Therefore, it is critical to develop new strategies to eliminate cancer cells, especially those with metastatic potential. In this regard, gene therapy provides great opportunities to effectively eradicate cancerous cells. SALL4 is one of the gene-therapy candidates involved in the progression of various cancers.
SALL4 belongs to the SALL gene family and encodes a zinc-finger transcription factor. As a mediator of tumorigenesis process, this gene has been introduced as a new diagnostic indicator for early detection of several cancers including BC 5-7. SALL4 is important for stem cells to maintain their pluripotency and self-renewal capacities 8,9. Overexpressed SALL4 has been shown to regulate apoptosis, proliferation, tumor invasion, migration, and drug resistance in various malignancies 10. Several studies have shown that the inhibition of SALL4 expression in cancer cells by siRNA reduces their viability and impairs their invasiveness in vitro 11.
In today molecular medicine, siRNAs offer best candidates for specifically targeting and downregulating oncogenes. However, clinical application of siRNAs faces a major challenge, and that is their delivery to cancer cells 12. Non-viral vectors are excellent candidates for delivering siRNAs; however, these vectors have limitations regarding the sizes of genes to be carried, as well as issues such as immunogenicity, cytotoxicity, and insertional mutagenesis. Polymer-based non-viral gene carriers, on the other hand, present acceptable safety profile and relatively simple construction. Furthermore, they are less likely to promote immunogenicity, and can be repeatedly administered with little toxicity 13-15. Cationic phospholipids, inorganic nanoparticles, and polycations comprise nanocarrier systems for transferring nucleic acids into target cells 16-18. Polyethyleneimines (PEIs) are gold standard carriers in polycation-based transfection systems. PEIs with various molecular weights, structures, and chemical groups have been widely used in many in vitro and in vivo studies 19.
Recently, PEI-based polymeric vectors have been developed to augment the efficiency of target-specific intracellular gene delivery. Regarding that membrane receptors that mediate endocytosis are often over-expressed on tumor cells 20, these receptors can promote the internalization of endosomal vesicles. These early endosomes then transform to late endosomes before being merged with lysosomes 21. Low-density Lipoprotein-Related Protein 2 (LRP-2), which is also known as megalin, is a 600‐kDa transmembrane endocytic receptor mediating the internalization of various ligands such as apolipoproteins 22. This protein is particularly over-expressed on the surface of MCF-7 cells 4, so it has the potential to be used in receptor-mediated gene delivery systems along with modified PEIs for targeting genes into BC cells. The pH-buffering capacity of nano-polyplexes is a crucial feature of cationic polymers to suppress lysosomal degradation of biomolecules such as nucleic acids 23,24.
In this study, megalin-targeted amino-glycoside-carboxyalkyl-PEI 25 was used for transfecting SALL4- siRNA into MCF-7 cells (Figure 1).
Materials and Methods :
PEIs ligand (PEI25G10C50) preparation: In order to prepare PEI25G10C50, 6-bromohexa-noic acid which is a derivative of PEI was first synthesized and then coupled with aminoglycoside to form carboxyalkylated PEIs. Necessary investigations were carried out in accordance with our previous study 16.
Particle size: Dynamic Light Scattering (DLS 9900, K-One.Ltd, Korea) was used to measure mean hydrodynamic particle size and the size distribution of polymer/pDNA polyplexes. The C/P=2, C/P=4, and C/P=6 cationic polymers were diluted in 115, 105, and 95 µl Double-Distilled Water (DDW) and then in equal volumes of the same buffer containing 5 µl pDNA. Afterwards, the samples were diluted four times, and the results were presented as mean values.
Atomic force microscopy: Atomic force microscopy (JPK NanoWizard II, Germany) was used to investigate the surface properties, morphology, and sizes of polyplexes. For this, polymers and pDNA were diluted in Tris-HCl buffer (Tris-HCl 10 mM, NaCl 10 mM, MgCl2 2 mM). Then, 10 µl of each solution was placed on a slide, and after fixing, microscopic images of nanoparticles were examined.
Cell culture and transfection: MCF‐7 cell line was purchased from Pasteur Institute of Iran (Karaj, Iran). DMEM medium containing 10% Fetal Bovine Serum (FBS, Gibco, UK), 100 μg/ml streptomycin, and 100 IU/ml penicillin (Gibco, UK) was used to culture the cells at 37°C and 5% CO2. The transfection was conducted by SALL4 siRNA/shRNA/ RNAi Lentivector and Scrambled siRNA GFP Lentivector piLenti-siRNA-GFP plasmid (Blood Transfusion Research Center, Iran) 25. The required volumes of the synthesized carriers and plasmids were admixed with serum-free culture medium at Carrier-Plasmid (C/P) ratios of 2, 4, and 6. To form polyplexes, the mixture was incubated at room temperature (20 min). Four hr after the transfection, the old medium was replaced with DMEM supplemented with 10% FBS, and 1% penicillin-streptomycin at 37°C and 5% CO2. The sequence of SALL-4 siRNA was as 5´-TCTGAGTTC CTGGAACATAAGAAAAATTG-3´, and that of the scrambled siRNA (Negative control) was as 5´-GGGTGAACTCACGTCAGAA-3´.
Cytotoxicity measurement: The MTT method was applied to assess the cytotoxicity of the polyplexes against 1×104 MCF-7 cells seeded in 96-well plates containing 100 ml DMEM warm up supplemented with 10% FBS. After 24-hr incubation, 25 μl of the polyplexes (i.e. 200 ng plasmid) at the C/P ratios of 2, 4, and 6 was added to each well. After another 4-hr incubation at 37°C, the medium was substituted with 100 ml fresh serum (10% FBS). After 48 hr, the MTT reagent was added to the wells, and the plates were further incubated at 37°C for 3 hr. Then, the medium was discarded, and formazan crystals were dissolved by adding 100 μl of DMSO. The absorbance at 570 nm was finally read by a microplate reader (Crocodile 5-in-one ELISA mini Workstation, Titertek Berthold). The mean viability of the cells was calculated considering control cells as: A treated/A control × 100. The experiment was conducted in triplicate.
Qualitative assessment of transfection efficiency: MCF-7 cells (0.5×105) were cultured in 12-well plates and treated (For 4 hr) with the C/P4 and 6 nano- polyplexes prepared by SALL4 siRNA/shRNA/RNAi Lentivector, Scrambled siRNA GFP Lentivector piLenti-siRNA-GFP plasmid. After 72 hr of the transfection, GFP expressing cells were observed by fluorescence microscopy (Olympus Bx51, Japan).
Quantitative assessment of transfection efficiency: To assess the efficiency of the transfection, the expression of GFP was determined using flow cytometry (BD Biosciences, USA). MCF-7 cells were transfected by the vectors at the C/P ratios of 2, 4, and 6. After 72 hr, the transfected cells were detached by adding 120 µl trypsin-EDTA, which was deactivated by adding 300 µl cold PBS. Then, 1 µl of propidium iodide (1 mg/ml) was added to the samples. The cell suspension was directly assessed via a flow cytometer equipped with a 488 nm xenon-ion excitation lamp. Data analysis was conducted using the FlowJo software.
RNA extraction and real-time PCR: First, RNA was extracted (Favorgen Biotech Corp., Taiwan), and its purity was assessed by spectrophotometry (Lambda max, Japan) and reading the absorbance ratio of 260/280 nm. After that, cDNA was synthesized using PrimeScript Reverse Transcript Reagent Kit (GenetBio, Korea). Quantitative SYBR green (Favorgen Biotech Corp., Taiwan) real-time PCR (qRT-PCR) was performed (Rotor-Gene 6000, Qiagen, Germany) to determine the mRNA expression of SALL4, applying appropriate primers (Table 1). Using the mRNA level of GAPDH, the relative concentration of SALL4 mRNA was determined and adjusted using the 2−ΔΔCT method. The experiment was conducted in duplicate.
Scratch assay: After the initial seeding of 0.1×106 MCF-7 cells in 12-well plates and reaching a confluency of 70%, the transfection was performed using the C/P ratio of 6 for SALL4 siRNA/shRNA/RNAi Lentivector and Scrambled siRNA GFP Lentivector piLenti-siRNA-GFP plasmid as previously described. After 48 hr of the transfection, a plastic tip (0.1 mm) was utilized to scratch cellular monolayers. To eliminate cell debris, the wounded monolayers were double-washed with PBS. Inverted microscopy was used to determine the distance of cell migration towards the damaged surface at 0 and 24 hr after scratching. The images were analyzed by the MRI-Wound-Healing Tool plugin of the ImageJ software.
Statistical analysis: Mean±SD values were calculated based on duplicate or triplicate experiments. The differences between groups were assessed by ANOVA following Tukey's post-hoc test. SPSS v.25.0 (SPSS, Inc., Chicago, IL, USA) was utilized for statistical analysis. Graphs were drawn by GraphPad Prism 8.0.2 (GraphPad Software, Inc., La Jolla, CA, USA). Statistical significance was assigned as p<0.05.
Results :
Particle size and zeta potential of the modified PEIs: To be effective, polymeric gene delivery systems need to have ideal charge density and particle size, and in fact, these are significant factors influencing the cellular uptake of gene carriers. Previous studies have suggested that the appropriate size of polyplexes for gene delivery is approximately 30-500 nm 26. Polymeric gene delivery systems have been subjected to a variety of modifications of their physical and chemical properties to improve transfection efficiency 16,27. So far, new methods have been developed to produce NPs with optimal size distribution 28. Compared with non-modified PEIs in our previous report, we here succeeded producing relatively small-size nanoplexes using aminoglycoside-carboxyalkylated PEIs.
In this study, the mean hydrodynamic diameter of PEI/plasmid complexes was determined by DLS (DLS 9900, K-One.Ltd, Korea). The average sizes of PEI/plasmid complexes in C/P=2, C/P=4, and C/P=6 conditions were 144, 244, and 367 nm, respectively. Intensity distribution graph also showed that the both C/P ratios of 4 and 6 were uniform in size (Figure 2A-C).
Size distribution of modified PEIs based on atomic force microscopy (AFM): AFM micrographs showed that the nanoparticles produced by the scramble plasmid and PEI25G10C50 were spherical and uniform with sizes of 145-160 nm (Figure 2D).
The cytotoxicity of modified PEIs against MCF7 cells: The cytotoxicity of the modified PEIs against MCF7 cells was assessed at the 4 and 6 C/P ratios of the nano polyplexes prepared with Scramble siRNA GFP Lentivector piLenti-siRNA-GFP plasmid. Forty-eight hr after the transfection, the viability of MCF7 cells was measured using MTT assay. The results showed that the both concentrations of the polyplexes had almost no toxicity against MCF-7 cells (cell viability of 95-100%, Figure 3A).
Qualitative assessment of GFP transfection by fluorescence microscopy: SALL4 siRNA/shRNA/RNAi Lentivector and Scramble siRNA GFP Lentivector piLenti-siRNA-GFP plasmid were used to qualitatively assess the efficiency of in vitro transfection of the modified PEIs (C/P ratios of 4 and 6) into MCF-7 cells. Accordingly, the transfection efficiency was higher in the C/P=6 compared to C/P=4 (Figure 3B).
Analysis of GFP expression: The transfection efficiencies of the GFP reporter gene in MCF-7 cells at the C/P ratios of 2, 4, and 6 of the polyplex were evaluated by flow cytometry (BD Biosciences, USA). The results showed that the transfection efficacy increased at C/P=6 compared to C/P=4 and C/P=2 (Figure 4A). Transfection efficacies at C/P=6 in the scramble and siRNA transfected cells were 35.9 and 29.12%, respectively. Therefore, our data showed that the transfection efficiency was enhanced by increasing the polymer concentration.
Transfection of SALL4- siRNA significantly reduced SALL4 gene expression: The SALL4 expression significantly reduced in the cells transfected with SALL4-siRNA compared to those transfected with Scramble (8-fold) and untreated cells (14-fold, Figure 4B).
Decreased SALL4 expression reduced migration of MCF-7 cells: In this study, after silencing SALL4 by siRNA and PEI25C50G10, wound healing assay was carried out, and images were prepared at 0 and 24 hr after the transfection (Figure 5A). The results showed that using the C/P=6 concentration of the nano-polyplex decreased the migration of MCF-7 cells by 58% (Figure 5B). According to figure 5A, differences in scratch areas (analyzed by ImageJ software) between the groups transfected with SiRNA-SALL4, Scramble, and Untreated cells after 24 hr indicate a reduction in the migration of MCF-7 cells transfected with SiRNA-SALL4 compared to other groups. The above sentences were added to the manuscript.
Discussion :
Breast cancer as the most malignancy in women is one of the research priorities for new diagnosis and treatment, gene therapy particularly. One of the methods of gene therapy has substaintially developed in cancer years, using siRNA technology to regulate gene expression involved in cancer progression. However, targeted delivery is still the most challenging issue in this strategy. In this regard, numerous non-viral vectors including cationic polymers have been employed to target specific genes in cancer cells 29.
PEIs are the most frequently used polycations to target siRNA in to cells. Two important advantages of PEIs are the stability of toroidal polyplex particles and the target buffering potential at almost any PH, which augment their effectiveness in the proton sponge process. Moreover, PEIs can be modified with various functionalized groups to enhance their targeting efficiency and reduce their cytotoxicity 30,31.
So far, some studies have applied modified PEI-based polymers to target nucleic acids in to megalin expressing cells and to increase the efficiency of siRNA delivery systems. Xue et al synthesized a PEI polymer using apolipoprotein E as a ligand of megalin. In the recent report, using Ap-LPNs rather than uncoated LPNs increased the efficiency of the transfection of anti clustrin siRNA in to chmo-resistant MCF7-cells three folds, which resulted in clustrin suppression and chemosensitization to paclitaxel in cancer cells 4.
Another modified PEIs were recently developed to target nucleic acids in to megalin expressing cells consisted of aminoglycoside-carboxyalkylpolyethyleni-mine25 covalently bound to aminoglycosides (Gentamicin) that used in the current study. In the previous research have shown that high C/P ratios of aminoglycoside carboxyalkylpolyethylenimine25 is highly toxic against kidney cells 16. Therefore, C/P ratio >6 was not used in this study and MTT results were confirmed ultralow cytotoxicity C/P6 in MCF7 cells. So, in the present study, glycoside-carboxyalkyl-PEI25 was targeted by gentamicin, used for transfection of siRNA-SALL4 to MCF7 cells as megalin expressing cell line.
SALL4 oncogene, through modulating various signaling pathways, participates in the proliferation, apoptosis, migration, and invasion of several types of malignant cells. For example, the effect of SALL4 silencing on MCF-7 cells apoptosis was recently investigated and it was demonstrated that SALL4 knock down was associated with reduced expression of Bcl2 and elevated apoptosis 25. Furthermore, SALL4 has been shown to be important in maintaining the mobility and migration of tumor cells in the neoplasms of lung, stomach, and breast. Itou et al showed that SALL4 was involved in shifting the basal cell morphology from spindle-shaped to spherical and enhancing cellular migration by regulating α6β1 integrin expression and modulating PI function 32. Moreover, SALL4 augmented cellular migration by inhibiting PETEN expression and subsequent blocking of the AKT/mTOR signaling pathway 33. The result of these studies aligned with conclusions of MTT assay in our study, so that down regulate of SALL4 lead to significantly declined of MCF7 cells migration.
On the other hand, Jiang and Wang who showed that silencing SALL4 with miR-16 reduced the migration and proliferation of gastric cancer cells 34. This and similar studies have confirmed same effect SALL4 in breast cancer and other malignancies.
Therefore, we believe the knockdown of SALL4 reported in this study with glycoside-carboxyalkyl-PEI carrier use for controlling of migration in various cancer.
Conclusion :
In this study, polycationic aminoglycoside-conjugat-ed carboxyalkylated-PEI was used as a non-viral vector to deliver siRNA-SALL4 into MCF-7 cells. Megalin was exploited as one of the most important receptors on MCF-7 cells to mediate endocytosis. According to our results, using the modified PEIs increased the transfection efficiency. Furthermore, silencing SALL4 significantly suppressed the migration of MCF-7 cells. It is suggested to consider PEI25G10C50 as an efficient gene-delivery system to target SALL4 gene towards cancer cells in patients with BC.
Acknowledgement :
This project was funded by North Khorasan University of Medical Sciences, Bojnurd, Iran (Grant no. 970036). Ethics approval code is IR.NKUMS.REC. 1397.081.
Conflict of Interest :
The authors declare that they have no conflict of interest.
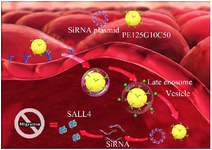
Figure 1. The study protocol. 1) The PEI25G10C50 integrates with plasmid and binds to megalin on the surface of MCF-7 cells. 2) Nano-polyplexes enter into the cells through receptor-dependent endocytosis. 3) Plasmid and PEI25G10C50 are released into the cytoplasm by the endosomal escape process at late endosome stage. 4) The siRNA degrades SALL4 mRNA.
|
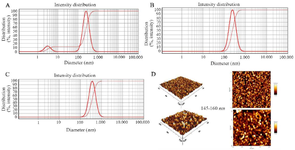
Figure 2. The intensity size distribution of PEI25G10C50. A) C/P=2; B) C/P=4; C) C/P=6. D: Atomic force microscopy micrographs of polyplexes formed with Scrambled siRNA GFP Lentivector piLenti-siRNA-GFP plasmid and PEI25G10C50.
|
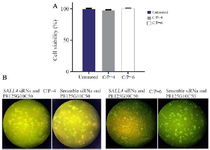
Figure 3. A) The cytotoxicity of polyplexes prepared with Scrambled siRNA GFP Lentivector piLenti-siRNA-GFP plasmid and PEI25G 10C50 against MCF-7 breast cancer cell line. Means±SD from 3 repeats have been presented. B) Fluorescent imaging of GFP expression in MCF-7 cells transfected with the polyplexes prepared with GFP-expressing plasmid DNA and PEI25G10C50. Photomicrographs show green fluorescence expression in MCF7 cells at different C/Ps concentrations, 72 hr after the transfection.
|
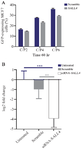
Figure 4. A) Transfection efficiency using the polyplexes prepared with GFP-expressing plasmid (SALL4 siRNA and scramble siRNA) and PEI25G10C50 at different C/P concentrations. The number of GFP-expressing cells was determined as the mean value from 3 measurements. B) The relative expression of SALL4 in untreated, scrambled, and siRNA-SALL4 transfected MCF-7 cells. The expression of SALL4 significantly decreased in the cells transfected with SALL4- siRNA compared to either the cells transfected with scrambled or untreated cells. The SALL4 expression in the scrambled group was not significantly different compared to untreated group. The data was normalized considering the untreated control group (log2). n=3 or 4. **: p<0.01, ***: p<0.001.
|
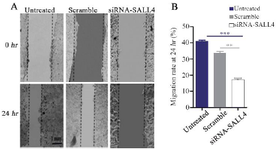
Figure 5. Assessment of MCF-7 cells migration by the scratch assay. Silencing SALL4 significantly decreased the migration of MCF-7 cells in comparison with untreated and scrambled transfected cells. The results have been expressed as mean±SD (3 repeats) of migration after 24 hr of transfection compared to baseline**: p<0.01.
|

Table 1. The sequences of the primers used for SALL4 and GAPDH
* Primers were designed to contain two SALL4 isoforms.
|
|