Distribution of Class I Integron and smqnr Resistance Gene Among Stenotrophomonas maltophilia Isolated from Clinical Samples in Iran
-
Tabaraie, Bahman
-
Kousha Faravar Giti, Industrial Research Institute of Biotechnology, Tehran, Iran
-
 Afrough, Parviz
Pasteur Institute of Iran, Tehran, Iran, Tel: +98 21 66953311, Fax: +98 21 66492619, E-mail: afroughparviz9@gmail.com
Afrough, Parviz
Pasteur Institute of Iran, Tehran, Iran, Tel: +98 21 66953311, Fax: +98 21 66492619, E-mail: afroughparviz9@gmail.com
-
Microbiology Research Center, Pasteur Institute of Iran, Tehran, Iran
-
Department of Mycobacteriology and Pulmonary Research, Pasteur Institute, Tehran, Iran
-
Behrouzi, Ava
-
Department of Mycobacteriology and Pulmonary Research, Pasteur Institute, Tehran, Iran
Abstract: Background: Stenotrophomonas maltophilia (S. maltophilia) is a multiple-antibiotic-resistant opportunistic pathogen that is being isolated with increasing frequency from patients with health-care-associated infections. S. maltophilia is inherently resistant to most of the available antimicrobial agents. Spread of resistant strains has been attributed, in part, to class I integrons. In vitro susceptibility studies have shown trimethoprim-sulfamethoxazole and new floroquinolones as two important agents with activity against these organisms.
Methods: 150 isolates of S. maltophilia were isolated from clinical samples such as respiratory discharges, sputum, and catheter and hospital environments. These isolates were also subjected to susceptibility testing and polymerase chain reaction for four groups of genes including int encoding integron elements, sulI and sulII encoding trimethoprim-sulfamethoxazole resistance and smqnr encoding quinolone resistance.
Results: The rate of resistance to trimethoprim-sulfamethoxazole was up to 27 (18%) and the highest resistance to quinolone family belonged to ofloxacin (20%) and the lowest rate was for gatifloxacin (16%). The results showed that 14% of isolates contained integron elements concomitantly with sulI and sulII genes.
Conclusion: Resistance rate of S. maltophilia to co-trimoxazole and fluoroquinolones and detection of integron elements between isolates in this study showed that this rate corresponded to other data obtained from other studies.
Introduction :
Stenotrophomonas maltophilia (S. maltophilia) is an aerobic, nonfermentative, gram-negative, catalase-positive and oxidase-negative bacterium. S. maltophilia is ubiquitous in aqueous environments including water, urine, or respiratory secretions, soil and plants 1. This bacterium causes nosocomial infections in immunocompromised patients and frequently colonizes breathing tubes such as endotracheal or tracheostomy tubes, the respiratory tract and urinary catheters. Infection easily commences by the presence of prosthetic material (plastic or metal), and the most effective treatment is removal of the prosthetic devices. Therefore, growth of S. maltophiliaisolated from respiratory or urinary specimens in microbiological media is difficult to interpret and not a proof of infection. However, isolation of S. maltophilia from sterile body regions (e.g., blood) usually represents true infection. In immunocompetent individuals, S. maltophilia is a relatively unusual cause of pneumonia, urinary tract infection, or blood stream infection. S. maltophilia is naturally resistant to many antibiotics (including all carbapenems) and often difficult to eradicate. Although resistance has been increasing, many strains of S. maltophilia are sensitive to co-trimoxazole and ticarcillin 2. It is not usually sensitive to piperacillin, and sensitivity to ceftazidime is variable. S. maltophilia is resistant to many β-lactams, β-lactamase inhibitors, and aminoglycosides 3,4.
A recent survey has indicated that newer fluoroquinolones demonstrated good efficacy against these bacteria. The most active antimicrobials were levofloxacin and gatifloxacin in which resistance rates were reported to be 6.5 and 14.1%, respectively 5. Because of low resistance levels (~5%), trimethoprim-sulfamethoxazole (TMP/SMX) has remained the choice of antimicrobial therapy against S. maltophilia infections worldwide. Although there are a few surveillance studies for Stenotrophomonas infections, resistance to TMP/SMX appears to be emerging, and recent in vitro modeling studies have shown that combination therapies by TMP/SMX plus ciprofloxacin or TMP/SMX plus tobramycin exhibit a greater killing capacity than TMP/SMX alone 2,6. In addition, S. maltophilia can acquire antimicrobial resistance through integrons, transposons, and plasmids. Class 1 integrons have been characterized from S. maltophilia strains isolated in Argentina and Taiwan, which indicates that they contribute to TMP/SMX resistance through the sulI gene carried as part of the 3´ end of the class 1 integron 7. Sul genes have been reported to contain class 1 integrons and insertion element common region (ISCR) elements that are responsible for high rate of resistance to TMP-SMX in S. maltophilia 8,9.
Recent analysis of S. maltophilia has identified a novel family of resistance genes (smqnr) encoding proteins containing pentapeptide repeats, which confer low-level resistance to quinolones 10. Qnr gene exists in several bacterial genera and is located in S. maltophilia chromosome, designated as smqnr which encoded a protein that contributes to intrinsic resistance to quinolones 9. This gene could be plasmid-borne and results in high resistance to quinolones in wild type and mutant bacteria 11,12.
Examination of K279a and another S. maltophilia genome sequence (R551-3) also identified a novel family of genes (smqnr) that encode proteins with homology to the Qnr quinolone protection proteins found in the Enterobacteriaceae 5. These Smqnr proteins (QnrA, B and S) in which pentapeptide repeats could be found, confer low-level quinolone resistance by protecting DNA gyrase and topoisomerases. In Enterobacteriaceae members, Smqnr proteins are usually located in association with other resistance determinants on large plasmids 13. The clinical importance of plasmid-mediated quinolone resistance is uncertain, although it is postulated that it may help stabilize or select for mutations in the Quinolone Resistance-Determining Region (QRDR) of DNA gyrase and topoisomerase, which then confers high-level quinolone resistance 13. This study could determine the relation between dissemination and increasing antibiotic resistance of S. maltophilia isolated from hospital and patient's samples.
Some studies have been performed in Iran that focused on S. maltophilia antibiotic resistance. In one typical study, 3% of 895 isolates were resistant to co-trimoxazole.
Materials and Methods :
Bacterial strains: During a two year period between 2012 to 2014, 150 isolates of S. maltophilia were collected from different clinical settings in Tehran, Iran and clinical samples like respiratory samples, ventilator associated pneumonia, discharges of patients, surgery devices and catheters. Body fluids were inoculated to BACTEC media. BACTEC system was pereferred for better performance in growth and identification of microbial agents because detection procedure was based on on-time and precise computed mechanism.
Antibiotic susceptibility testing: Susceptibility testing was performed by disc diffusion methods (Mast diagnostics) and E-test (AB Biodisk) for trimethoprim-sulfamethoxazole, ciprofloxacin, ofloxacin, gatifloxacin and moxifloxacin on Muller Hinton agar as described by Clinical and Laboratory Standards Institute. Resistant strains to trimethoprim-sulfamethoxazole and quinolones were stored at -70oC.
Polymerase chain reaction amplifications: Extraction and purification of DNA from bacterial colonies and plasmids was accomplished by commercial extraction kits from the isolates (QIAmp mini kit from Qiagen). Polymerase Chain Reaction (PCR) was carried out in 50 ml containing 2 μ l template DNA, 5 μ l 10x concentrated PCR buffer, 1 μl of each appropriate primer, 10 μl dNTPs, 1 μl Taq DNA polymerase, and 21 μl sterilized distilled water 10. Primer designation and sequences are shown in table 1.
Results :
Antimicrobial susceptibility testing: Strains were tested for susceptibility to trimethoprim-sulfamethoxazole, ciprofloxacin, ofloxacin, gatifloxacin and moxifloxacin. The rate of resistance to TMP/SMX was up to 27 (18%) and this rate for quinolone family was: ciprofloxacin 27 (18%), gatifloxacin 24 (16%), moxifloxacin 25 (17%), and ofloxacin 30 (20%). The TMP/SMX -resistant isolates possessed MICs >32 μg/ml, whereas the sensitive controls possessed TMP/SMX MICs ranging from 0.5 to 2 μg/ml. MIC for ciprofloxacin resistant strains was >4 μg/ml and for gatifloxacin resistant strains, moxifloxacin resistant strains and ofloxacin resistant strains of S. maltophilia was >8 μg/ml (Table 2).
Distribution of class I integron: 14% of strains which were resistant to TMP/SMX contained integron class 1 using primers int I F, int IR (5´ conserved region) with DNA bands of 510 bp (Figure 1). Out of the 27 TMP/SMX -resistant S. maltophilia isolates analyzed, 7 isolates possessed the sulI gene. SulI gene was located as part of the 3´ end of a class 1 integron. None of the trimethoprim-sulfamethoxazole -susceptible S. maltophilia isolates yielded positive sulI PCR products. Out of 27 TMP/SMX -resistant S. maltophilia isolates, 12 isolates carried sulII gene using sulIIF and sulIIR primers (Figure 2). None of the TMP/SMX-susceptible S. maltophilia isolates displayed positive sulII PCR products. Of the 27 TMP/SMX -resistant S. maltophilia isolates, 5 strains concomitantly contained sulI and sulII genes. Out of the 106 resistant S. maltophilia isolates,16 (10%) isolates contained smqnr genes (Figure 1).
Discussion :
In this study, two groups of antibiotics including trimethoprim-sulfamethoxazole and some of quinolone members such as ciprofloxacin, gatifloxacin, moxifloxacin and ofloxacin were selected as well-known antimicrobial agents against S. maltophilia. An attempt was made to analyze genetic determinants responsible for drug susceptibility pattern of 150 S. maltophilia strains isolated during a 6 month period in 2010 collected from different places of hospital and clinical samples like blood and respiratory samples, ventilator associated pneumonia, discharges of patients, surgery devices and catheters. The results showed that the resistance of these S. maltophilia isolates to TMP/SMX and quinolone has slightly risen. By comparison in our study, it can be concluded that 44% of SXT resistant isolates contained large plasmids including sulI genes.
Out of these SXT resistant isolates, 27% of them carried sulI gene in class 1 integron. These results implied that most of SXT resistant strains contain plasmids for SXT resistance. This plasmid could usually be observed between Enterobacteriaceae members. Most studies of the location and dissemination of sulII genes have concentrated on Enterobacteriaceae, such as Escherichia coli (E. coli) and Salmonella enteric 8. The data presented in this study showed that sulII gene may spread by Enterobacteriaceae origins among S. maltophilia strains. These data suggest that microbiology laboratories need to carefully monitor S. maltophilia strains which show resistance to TMP/SMX, because they have the potential to increase by means of mobile genetic elements.
Betrieu et al showed that 91% of strains were susceptible to SXT and MIC 14. Based on a study in 2001, rates of resistance to SXT ranged from 2% in Canada and Latin America to 10% in Europe 15. In another study performed in Saudi Arabia in 2006, two resistant cases to SXT were reported. Both isolates were resistant to TMP-SXT (MIC >8/152 μg/ml by MicroScan system and MIC >32 μg/ml by E-test strip). The two isolates were also resistant to gentamicin (MIC >8 μg/ml), both meropenem and imipenem (MIC >16 μg/ml) and ciprofloxacin (MIC >4 μg/ml). They were sensitive to ceftazidime (MIC <2 μg/ml) and ticarcillin-clavulanate (MIC=16/2 μg/ml). The sensitivities to amikacin, chloramphenicol, tetracycline, levofloxacin, aztreonam and piperacillin- tazobactam were variable between the two isolates 2. In a study performed in England in 2005, it was indicated that none of S. maltophilia isolates from salad was resistant to trimethoprim-sulfamethoxazole, ciprofloxacin, but these isolates concomitantly were resistant to chloramphenicol; 7 (78%) to piperacillin/tazobactam; 5 (56%) to ceftazidime, and 2 (22%) to gentamicin. But it should be considered that the number of samples (salad) in the mentioned study was so small and cannot be compared with this study because our study was about clinical samples and there were many specimens 16.
It can be concluded that as the time passes, the rate of resistance of first line effective antibiotics to S. maltophilia developes and many isolates should be considered for testing in laboratory. The most significant study ever performed on susceptibility of S. maltophilia was a study in 1999 in Department of Microbiology and Immunology, Queen’s University, Kingston, Ontario K7L 3N6, Canada entitled "Multiple Antibiotic Resistance in Stenotrophomonas maltophilia: Involvement of a Multidrug Efflux System".
Conclusion :
In this study, the mechanisms of resistance and percentage of susceptibilities to antibiotics were indicated 4. There are some studies performed in Iran which focused on S. maltophilia isolates and its antibiotic resistance. In a study in 2011 among a total of 12922 blood specimens, 2300 specimens had a positive blood culture (17.7%); the specimens were collected early at hospitalization and as a result, blood samples were collected before initiation of any treatment. Not considering fungal growth, 21 microorganisms were recognized, with S. maltophilia being the most common one (895 specimens; 38.9%). There were 95 sensitive and 5 resistant species in both the disk diffusion method and E-test for co-trimoxazole 25.
Acknowledgement :
The authors appreciate of laboratory team of Pasteur institute of Iran.
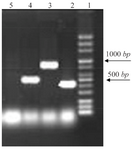
Figure 1. PCR results from right: lane 2, (sulI, 420 bp); lane 3 (smqnr 817 bp); lane 4 (int, 510 bp); lane 5 negative control.
|
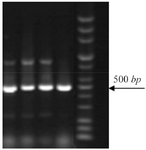
Figure 2. PCR amplification of sulII genes (450 bp).
|
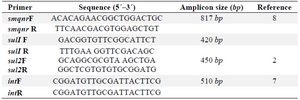
Table 1. List of primers used in this study
|
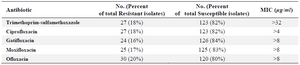
Table 2. Antimicrobial susceptibility testing results
|
|