Development of a Single Stranded DNA Aptamer as a Molecular Probe for LNCap Cells Using Cell-SELEX
-
Almasi, Faezeh
-
Department of Biotechnology, College of Science, Tehran University, Tehran, Iran
-
 Mousavi Gargari, Seyed Latif
Biology Department, Shahed University, Tehran-Qom Express way, Tehran, Iran, Tel: +98 21 51212200, E-mail: slmousavi@shahed.ac.ir
Mousavi Gargari, Seyed Latif
Biology Department, Shahed University, Tehran-Qom Express way, Tehran, Iran, Tel: +98 21 51212200, E-mail: slmousavi@shahed.ac.ir
-
Department of Biology, Shahed University, Tehran, Iran
Abstract: Background: Nowadays, highly specific aptamers generated by cell SELEX technology (systematic evolution of ligands by exponential enrichment) are being applied for early detection of cancer cells. Prostate Specific Membrane Antigen (PSMA), over expressed in prostate cancer, is a highly specific marker and therefore can be used for diagnosis of the prostate cancer cells. The aim of the present study was to select single-stranded DNA aptamers against LNCap cells highly expressing PSMA, using cell–SELEX method which can be used as a diagnostic tool for the detection of prostate cancer cells.
Methods: After 10 rounds of cell-SELEX, DNA aptamers were isolated against PSMA using LNCaP cells as a target and PC-3 cell lines for counter SELEX. Five DNA aptamers with more than 70% affinity were selected up on flow cytometry analysis of positive clones.
Results: Dissociation constants of two selected sequences (A12-B1) were estimated in the range of 33.78±3.77 and 57.49±2.214 pmol, respectively. Conserved secondary structures of A12 and B1 sequences suggest the necessity of these structures for binding with high affinity to native PSMA. Comparison of the secondary structures of our isolated aptamers and aptamer A10 obtained by protein SELEX showed similar stem-loop structures which could be responsible for the recognition of PSMA on LNCap cell surface.
Conclusion: Our results indicated that selected aptamers may turn out to be ideal candidates for the development of a detection tool and also can be used in targeted drug delivery for future smart drugs.
Introduction :
Cancer is the major public health problem around the world. According to The World Cancer Report, more than 60% of the world's cases and about 70% of the world's cancer deaths occurred in Africa, Asia, and Central and South America 1. Prostate cancer is the second most common cause of cancer death in US men. In 2015, an estimated 220, 800 men will be diagnosed with prostate cancer, and there will be 27,540 prostate cancer-related deaths 2.
Prostate-Specific Antigen (PSA) was considered to be the best tumor marker for Prostate Cancer (PCa); however, the major drawbacks of PSA are inability in predicting metastatic potential and differentiating Benign Prostatic Hyperplasia (BPH) from PCa. Recently, a single and highly specific prostate trans-membrane glycoprotein has been identified to fill in the diagnostic gaps 3,4. Expression of Prostate-Specific Membrane Antigen (PSMA) is highly restricted and up regulated in advanced carcinoma. This marker seems to be inversely regulated in prostate cells 5. In many cases, diagnosis of cancer often comes after it has metastasized throughout the body, thereby early diagnosis can decrease the prostate cancer specific mortality rate. Numerous technical efforts have thus been made over the past decade to reliably detect cancer cells. Nucleic acid aptamers have been proposed as a new potential solution to overcome many of the drawbacks of current detection assays. Aptamers are synthetic oligonucleotide ligands with affinities and specificities for their target comparable to those of antibody/antigen interactions. Aptamers have been described for recognizing a wide variety of targets, from small chemical compounds to large multi-domain proteins and whole cells. Comparable attractive features of aptamers against antibodies, such as short generation time, fast tissue penetration, easily modification, long-term stability and low immunogenicity have made aptamers excellent molecular probes for clinical and therapeutic applications such as targeted therapy, detection and diagnostics 6.
Cell-SELEX is a modification of the traditional SELEX process targeting whole living cells. Aptamers developed by cell-SELEX are capable of binding to the native folding structures and glycosylation states of ligands without purifying or immobilization 7,8. The advantage of cell-SELEX-based aptamers especially in cancer research over protein SELEX, is to facilitate the development of molecular probes for diseased cell recognition. There have been several reports on cell-SELEX-based selection of aptamers against cancer cells 9. 2'-F-modified RNA aptamer (A10) binding to extracellular domain of PSMA was developed by protein SELEX 10. Conjugation of A10 to siRNAs and shRNAs 11,12, linked to gelonin 13, different anticancer drugs and nanoparticles 14-16 on prostate cancer cells have been studied.
In this research, due to simplicity of SELEX procedure with DNA and stability of DNA aptamers compared to RNA aptamers 17 and problems existing with highly glycosylated PSMA expression for doing protein SELEX, the cell-SELEX method was performed for selection of single-stranded DNA aptamers against LNCap cells which can be used in a diagnostic tool for the detection of prostate cancer cells.
Materials and Methods :
Chemicals and reagents: TA cloning vector and plasmid purification kits were obtained from Intron (Seoul, Republic of Korea). Lambda exonuclease was purchased from Thermofisher SCIENTIFIC (Canada). Yeast tRNA was purchased from Sigma (USA). Culture dishes were obtained from Nest (China). Unless otherwise noted, all chemicals were purchased From Merck (Germany).
Cell culture: Prostate cancer cell lines, LNCaP and PC-3, were purchased from the Pasteur Institute of Iran (Tehran, Iran) and were used for cell-SELEX. Cells were cultured in RPMI-1640, Gibco (USA) supplemented with 12% heat-inactivated Fetal Bovine Serum (FBS) (PAA-The Cell Culture Company) and 100 units/ml penicillin-streptomycin sigma (USA). All cultures were incubated at 37°C under 7% CO2 atmosphere.
Oligodeoxynucleotide library and primers: 80-mer synthetic DNA library containing a randomized 40 nucleotide with equal incorporation of A,G,C and T at each position flanked by primer binding sites CATCCATGGGATTCGTCGAC at 5´ and CTGCCTAGGCTCGAG at the 3´ was purchased from Daejeon (Korea). The 5′end labeled Fluorescein Isothiocyanate (FITC) forward primer and 3′end labeled phosphate reverse primer were obtained from Bioneer, Daejeon, (Korea). The FITC labeled primers were used for monitoring the progression of SELEX by flow cytometry (FACS cancytometern, Royan Institute, Tehran). The 3´ phosphorylated primers were used for the digestion of dsDNA by lambda exonuclease in order to get ssDNA.
Cell- SELEX procedure: PSMA positive LNCaP cells and PSMA negative PC-3 cells were counted and tested for viability before experiments. During SELEX procedure, cells were washed by washing buffer containing 4.5 g of glucose and 5 ml of 1 M MgCl2 in 1 L of Dulbecco's Phosphate-Buffered Saline (DPBS). The binding buffer used for selection was prepared by adding yeast tRNA and BSA into the washing buffer in order to reduce nonspecific binding.
To initiate in vitro selection, 5 nmol of the ssDNA pool was diluted in 1000 µl of binding buffer, denatured by heating at 95°C for 5 min and snap cooled on ice for 15 min. ssDNA was incubated with LNCaP cells (target cells) with continuous shaking on slow rocker for 1 hr at room temperature.
LNCaP cells were cultured into monolayers with at least 95% confluence in 100 mm×20 mm culture dish. Before each SELEX, cells were washed twice with washing buffer to eliminate dead cells and media components. After incubation, unbound sequences were discarded and 500 µl binding buffer (DNase-free water in the first round) was added to the washed cells. LNCaP bound sequences were recovered by cell scattering followed by heating cell-DNA complexes at 90°C for 10 min and centrifugation at 13000×g for 5 min. Supernatant containing eluted DNA was used as a template for PCR to obtain DNA pool for the next round of SELEX.
In order to eliminate the aptamers capable of binding both cells, counter SELEX (negative selection) was performed regularly on PC-3 cell lines after third SELEX. For negative SELEX, the bound sequences were directly incubated with PC-3 cells.
To select out the aptamers specifically recognizing PSMA, the stringency of the procedure was progressively increased by increasing the washing time and wash buffer volume, decreasing the incubation time from 1 hr to 15 min for target cell line and increasing from 15 to 60 min for PC-3cell line. Number of LNCaP cells was gradually reduced by culturing in 60 mm×15 mm culture dish from the third round and beyond. FBS was also gradually added to binding buffer from 10% in the eighth SELEX to about 20% in the last SELEX. After each SELEX, the DNAs bound to LNCaP or unbound to PC-3 were collected and amplified for the next round of selection.
PCR and aptamer strand separation: Aptamers resulting from each selection step were amplified by PCR in a volume of 50 µl with forward and phosphorylated reverse primers (25 cycles of 94°C for 30 s, 63°C for 30 s, 72°C for 30 s). Because of differences in amplification conditions between homogeneous DNA and aptamer libraries, the conditions were optimized for each round of Cell-SELEX. To optimize lambda exonuclease reaction, 5 µg of the purified dsDNA was incubated with 5 U lambda exonuclease in a total reaction volume of 30 µl in a reaction buffer at 37°C. The reaction was terminated after 0, 15, 25, 35, 45, 60 min of incubation, followed by 10 min incubation at 80°C and flash cooled on ice for 12 min.
Aliquots of digestion mixture at different incubation times were mixed with equal volume of RNA loading dye (8 M urea, 0.25% bromophenol blue, 1X TBE) and were analyzed on 10%, 8 M denaturing-urea PAGE (19 acryl-amide:1 bis-acrylamide, 8 M urea). ssDNA was extracted by ethanol precipitation method using 1/10th volume of 3 M NaOAC, 2 volumes of 100% ice-cold ethanol and 3 volumes of isopropanol and was quantified by picodrop (picodrop200, Biolab).
Flow cytometric analysis: In order to monitor the progression of selection process and enrichment of specific PSMA binders, fluorescence intensity of 4th, 7th, 9th and 10th SELEXs were measured in Royan Institute of Iran. Prior to monitoring, LNCaP and PC-3 cells were cultured overnight. Cells were dissociated by mild short time 1x trypsin treatment at room temperature. 50 pmol of ssDNA pool in 100 µl binding buffer was incubated with 5×105 cells of each cell lines which were resuspended in 100 µl binding buffer and 10% FBS for 45 min. Original FITC labeled library was used to adjust the fluorescence background.
Cloning, sequencing and structure determination: Amplified sequences of SELEX 10th with label free primers were cloned into pTG19-T vector. The cloned sequences were transformed into Escherichia coli (E. coli) BL21. Clones were analyzed with colony PCR to screen the positive clones. Plasmid DNA was purified from positive clones and sent for sequencing. Secondary structure determination of isolated aptamers and sequence analysis were performed by mfold software 3.8 and CLC sequence viewer 6.9.1, respectively.
Binding assay: Binding affinity of two aptamer candidates was determined using flow cytometry. 1×105 target cells were incubated with varying concentrations of FITC-labeled aptamer (0, 10, 50, 100, 200, and 300 pM) in a 200 μl volume of binding buffer containing 20% FBS. The FITC-labeled original library was used as a negative control to determine nonspecific binding. The equilibrium dissociation constant (Kd) of the aptamer-cell interaction was obtained by plotting the average total percentage of fluorescent cells (Y), against the concentration of aptamer (X) and the Bmax as the maximum number of binding sites, using a non-interacting binding sites model in SigmaPlot 12.0 according to Y=BmaxX/(Kd+X) equation (Jandel, San Rafel, CA, USA).
Results :
In vitro enrichment: Cell-SELEX was used to obtain DNA aptamers against PSMA positive cells. The schematic diagram of SELEX procedure is illustrated in figure 1. The binding of FITC-labeled ssDNA pools to both cell lines was measured after 4th, 7th, 9th and 10th rounds of selections.
Fluorescence signal of the target was significantly increased from 12.53% of the original library to 57.99% after 9th SELEX (Figure 2C). Additional rounds of SELEX didn’t show significance signal enhancement of the target. Inversely, the PC-3 cells showed great decrease in the fluorescence intensity and reached to minimum value by SELEX 10 (Figures 2B and 2D). Selection was completed at round 10, since no further significant signal difference was observed between last two successive selected pools. The highly enriched pools were cloned and binding affinity of 8 random positive clones was analyzed by flow cytometry. The results are exhibited in table 1. The binding affinity of clones A5, A12 and B1 to LNCaP cells was more than 75% compared to 12.53% of original library (Figure 3). The sequence of the highest binding aptamer (B1) is given in table 2.
Secondary structures of five selected aptamers were predicted using mfold software (Figure 4). dG of clones A12, B1, C10, A5 and B4 were -6.49, -7.85, -3.62, -5.92 and -5.03, respectively. The apparent dissociation constants (Kd) for the A12 and B1 were 33.78±3.77 and 57.49±2.214 pM, respectively (Figure 5).
Lambda exonuclease digestion: Results of dsDNA digestion by lambda exonuclease at different incubation times were analyzed on the 10% denaturing-urea PAGE (Figure 6). The band of double-stranded DNA disappeared gradually indicating the conversion to ssDNA up on 30 min of incubation time. Additional incubation times resulted in a decreased yield of ssDNA.
Discussion :
Although biological role of PSMA in PCa is unknown, correlation of cancer aggression and 100-1000 fold PSMA upregulation compared to the normal cells demonstrates its vital role in cancer spreading and intratumoral angiogenesis activity 18,19. PSMA expression has been related with aggressive disease in various circulating tumor cells and therefore can be an attractive option for detection and therapy of tumor cells.
Aptamer technology offers promising alternative options over antibodies for addressing the challenge of early diagnosis in human health and alleviates detection complications. For instance, aptamers can easily be forced to release the bound target without harsh treatments such as high heat, salt concentrations or pH, thus preserving the integrity of the target 20. Given the potential of aptamers for detection and targeted cell therapies, many efforts have been made over the past few years on optimizing selection methodologies for isolation of aptamers to cell-surface receptors. To date, several aptamers against receptors expressed on the surface of cancer cells have been identified using the cell-based selection approach. Cell surface SELEX can overcome the difficulties in obtaining purified preparations of recombinant membrane proteins for selection. In this research, successful generation of DNA aptamer was reported using cell- SELEX against LNCap cells.
Overexpression of PSMA even in poorly differentiated tumors is a better PCa indicator than the known PSA marker. Interestingly, its expression increases in higher-grade and metastatic prostate cancers while decreases in normal prostate, BPH and primary prostate cancer 18. As shown in figure 2, with increasing the number of selections, fluorescence intensity peak is shifted compared to the original library, indicating the ascending of high binding affinity aptamers within the pool during the SELEX. To prevent removal of probable target binding aptamers due to their low numbers at early steps of SELEX, counter SELEX was used after 3rd SELEX regularly. Counter-selection strategies eliminated aptamers capable of binding to similar surface proteins 21. In PSMA, highly glycosylated large extracellular domain accounts for more than 25% of the protein molecular weight in native form and this can be the main reason for preferring cell-SELEX rather than protein SELEX for obtaining DNA aptamers against PSMA although in whole cell SELEX technique, live cells are used as a target to isolate candidate aptamer without having any primary knowledge about the targets and their molecular properties. But PC-3 cell lines were used in counter SELEX to exclude the aptamers binding to the common antigen of LNCap and PC-3 cells. In the other way, the aptamers are directed toward the antigens which are over expressing and specifically present on the surface of LNCap cells, of which the PSMA is the most obvious one 18,19. Previously, it was demonstrated that, nanobody (VHH) produced against recombinant PSMA could efficiently attach to the surface of the LNCap cells whereas no immunoreactions were observed when applying anti-PSMA VHH on PC3 cells 22. This further supports our speculation that the developed aptamer could be against PSMA. Other researches have also used LNCap cells as PSMA positive and PC-3 as a PSMA negative cell line 23,24. Chang et al also demonstrated that different anti bodies produced against external domain of PSMA could recognize live and formalin fixed LNCap and PC3-PIP cell lines. None of the mAbs bound viable or formalin-fixed PC-3 cells are known to lack PSMA expression 25. Zamay et al used cell-SELEX for developing aptamers against cancer cells. DNA aptamers that bind to lung adenocarcinoma cells derived from postoperative patient tissues were selected 26. Negative selections were performed to remove aptamers that bind to healthy lung tissues in order to increase the specificity of the aptamers for lung cancer cells.
Despite the low nonspecific aptamer binding and easily controllable conditions during protein-SELEX, association of the integral proteins with cell matrix components, their assembly in lipid bilayer and post translational modifications 21,27 may affect the selected aptamers and they may not recognize the target in its physiological condition 28. Aptamers selected against recombinant targets have often failed postselection to bind to their targets on the surface of cells 29.
RNA aptamers selected for the histidine-tagged EGFRvIII ectodomain 30 and RNA aptamers for extracellular domain of human RTK Ret were unable to bind and recognize the native proteins 31. Recent progresses made in cell-based selection technologies have favored the isolation of aptamers specific for cell-surface proteins within their native milieu (i.e., the cell membrane). Generation of high-quality ssDNA is the most time consuming and essential critical step to ensure the success of SELEX 32. There are several drawbacks with some of ssDNA preparation methods such as generation of ssDNA with asymmetric PCR, loss of DNA tertiary structure due to fracture of bonds between biotin and streptavidin as a result of alkaline denaturation and presence of streptavidin as an additional target in biotin-streptavidin separation method and poor yield in size separation method 33,34. Lambda-exonuclease is a highly processive 5´to 3´enzyme acting in repair of breaks in the viral dsDNA 35. Despite the greatly reduced activity of the enzyme on single-stranded and non-phosphorylated DNA, the affinity of this exodeoxyribonuclease to phosphorylated 5'-end is 20 times higher than hydroxylated one (s) where ssDNA is produced in high yield and purity. Remaining bound to the DNA molecule until complete removal of the phosphorylated strand is another advantage of this enzyme in comparison to other exonucleases 32.
Citartan et al reported that the yield of ssDNA generation from pure dsDNA was about 40% more than unpurified PCR products 32. In the present work, purified PCR product was used for ssDNA preparation. Structural analysis of all high binding affinity aptamers after sequencing showed a conserved small stem loop exhibited with oval green in figure 4. There were also similar stem-loop structures in B4, A12 and B1 shown in red quasi-rectangle and similar hairpin in A5 and C10 shown in blue rectangle. Stem-loop structure in RNA aptamer against extracellular domain of PSMA (A10) closely similar to existing stem-loops displayed in green oval and blue rectangle in our work, have also been reported 35. Interestingly, aptamers, A12 and B1 with more than 78% affinity compared to library had conserved secondary structures (displayed in oval and rectangle shapes) suggesting the necessity of these two structures for binding with high affinity to native PSMA protein.
Conclusion :
Given the role of these aptamer targets in prostate cancer, these aptamers may turn out to be ideal candidates for the development of a detection tool and also can be used in targeted drug delivery for future smart drugs.
Acknowledgement :
The authors would like to thank the Biotechnology Development Council of I.R. Iran and Shahed University, Tehran, Iran, for supporting this study.
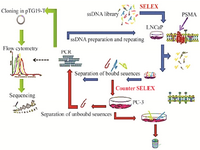
Figure 1. Schematic representation of the cell-SELEX.
|

Figure 2. Enrichment of selected DNA pools during SELEX monitored by flow cytometry. Assessment of the binding DNA pools to A) SELEX10, LNCaP; B) SELEX10, PC-3 cell lines. C) SELEX 9 LNCaP, D) SELEX 9 PC-3, E) comparison of SELEX4 and 7 LNCaP and F) comparison of SELEX4 and 7 PC-3.
|

Figure 3. The fluorescence intensity of 8 high binding affinity apta-mers to LNCaP cells. Affinity of A12 aptamer was 78.36% compared to 12.53% for original labeled library.
|
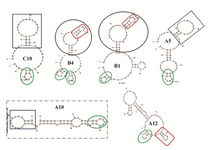
Figure 4. Secondary structure prediction. Secondary structures of five selected aptamers were predicted using mfold software. dG of aptamers A12,B1,C10, A5 and B4 were -6.49, -7.85, -3.62, -5.92 and -5.03, respectively. Structure inside the box is obtained from RNA aptamer selected against extracellular domain of PSMA.
|
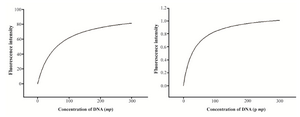
Figure 5. The equilibrium dissociation constant (Kd) of aptamers A12 and B1.
|
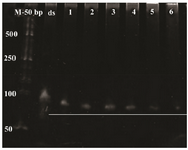
Figure 6. Urea PAGE for the time-course analysis of dsDNA apta-mer digestion. M: 50 bp ladder, ds: undigested double strand DNA, 1: DNA digestion for 15 min, 2: DNA digestion for 20 min, 3: DNA digestion for 30 min, 4: DNAdigestion for 40 min, 5: DNA digestion for 50 min, 6: DNA digestion for 60 min.
|
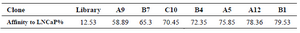
Table 1. Binding affinity of 8 random positive clones analyzed by flow cytometry
|

Table 2. Sequencing result of aptamer B only 40 variable nucleotides are given here. The two flanking known sequences are omitted
|
|