Outer Membrane Protein C (ompC) Gene as the Target for Diagnosis of Salmonella Species Isolated from Human and Animal Sources
-
 Al-Charrakh, Alaa H.
Department of Microbiology College of Medicine, Babylon University Hilla, Babylon Governorate, Iraq, Tel: +96 47813216822, E-mail: ahani67@gmail.com, aalcharrakh@yahoo.com
Al-Charrakh, Alaa H.
Department of Microbiology College of Medicine, Babylon University Hilla, Babylon Governorate, Iraq, Tel: +96 47813216822, E-mail: ahani67@gmail.com, aalcharrakh@yahoo.com
-
Department of Microbiology, College of Medicine, Babylon University, Hilla, Iraq
Abstract: Background: The use of selective and differential plating media is a simple method for the isolation of Salmonella spp. Recently, there has been a general move toward molecular methods of Salmonella detection and typing.
Methods: A total of 1200 different specimens collected from human and animal sources were involved in his study. 600 stool specimens from patients suffering from diarrhea and 600 specimens from gall bladder (bile) of cattle from Al-Diwaniya slaughter house, Iraq were used. Salmonella spp. were isolated and identified using bacterial culturing on selective media and colonies were tested by API 20Eand then serotyping through polyvalent antisera and conformation by Polymerase Chain Reaction (PCR). PCR was used to detect ompC gene encoding biosynthesis of outer membrane protein C of Salmonella genus.
Results: The results revealed that the rate of Salmonella isolates was 0.5% (3/600) from human and 1% (6/600) from animals. The PCR technique revealed that 9 isolates of Salmonella spp. harbored ompC gene. The results of this study revealed that the PCR technique had a high specificity in detection of Salmonella spp., in comparison to culture and biochemical test, Mini API 20 E and serological tests. The present study found no significant differences between human and animal isolates.
Conclusion: Detection of ompC gene is a good method for detection of Salmonella species isolated from clinical specimens. It has a high specificity in comparison with other tests, with its advantages of greater speed and effectiveness than conventional detection methods.
Introduction :
Enteric pathogens such as Salmonella enterica (S. enterica) cause significant morbidity and mortality. S. enterica serovars are a diverse group of pathogens that have evolved to survive in a wide range of environments and across multiple hosts 1. Infection begins with the ingestion of contaminated food or water so that salmonellae reach the intestinal epithelium and trigger gastrointestinal disease. In some patients, the infection spreads upon invasion of the intestinal epithelium, internalization within phagocytes, and subsequent dissemination 2. It has been estimated that there are more than 3 million deaths associated with Gram-negative enteric pathogens worldwide due to diarrhea and enteric fever each year 3. The use of selective and differential plating media is a simple method for the isolation of Salmonella spp. A wide variety of selective and differential media has been developed for this purpose, including xylose lysine desoxycholate agar (XLD), Hektoen Enteric (HE) agar, and bismuth sulfite (BS) agar 4. There has been a general move toward molecular methods of Salmonella detection and typing and PCR has become a potentially powerful alternative in microbiological diagnostics. While it is easy to amplify DNA derived from pure cultures, problems arise if the sample investigated is as complex as clinical specimen or food, since the PCR is easily inhibited by numerous substances, including humic acids, fats, and proteins. The ompC gene contains sequences unique to Salmonella isolates and demonstrates that this gene is a suitable PCR target for detection of Salmonella strains 5.
The aim of this study was to evaluate the rapid detection test to identify salmonellosis using the PCR technique for the detection of ompC gene of Salmonella genus, and to compare the PCR with traditional isolation and characterization methods currently used in diagnostic laboratories.
Materials and Methods :
Samples collection: A total of 1200 different specimens collected from human and animal sources were involved in this study, of which 600 stool specimens from patients suffering from diarrhea admitted to Al-Diwaniya Teaching Hospital for Maternity and Children (Al-Diwaniya province, Iraq) and 600 specimens from gall bladder (bile) of cattle from Al-Diwaniya slaughter house were collected. This study was conducted during the period that extended from May 2013 to April 2014.
From each specimen, 25 gm of human stool with a sterile wood spatula or animal bile were directly injected into an Erlenmeyer flask and 225 ml of buffered peptone water were added to obtain 1 part sample+9 part buffer, then mixed and incubated at 37°C overnight (16-20 hr). After that, 1 ml of the pre enrichment broth was transferred to 10 ml Tetrathionate broth and incubated at 37°C overnight (18-24 hr). Moreover, 0.1 ml (100 ul) of the pre-enrichment broth was also transferred to 10 ml Rappaport-Vassiliadis soy peptone (RVS) broth and incubated at 41°C overnight.
Isolation and identification of Salmonella isolates: For each specimen, a loopful (10 µl) from the incubated Tetrathionate broth (I) and RVS broth (II) was spread on XLD and on BGA agar plates and incubated at 37°C overnight (18-24 hr). For biochemical confirmation and serotyping, two suspected colonies (grown on XLD and BGA agar) were plated onto non-selective medium (nutrient agar) 6. Colonies that showed biochemical characteristics similar to that of Salmonella spp. were tested by Mini API 20E then serotyping. The identified isolates were then confirmed by PCR with ompC primers for detection of Salmonella genus 7.
DNA extraction and purification: One colony of each strain (cultured on solid medium) was inoculated into 5 ml of BHI (Broth Heart Infusion) and grown overnight at 37°C. From these cultures, DNA was purified from bacterial cells using Genomic DNA Mini kit (Geneaid, UK). Chromosomal DNAs obtained were used as templates for all PCR experiments. The PCR reactions were carried out in a thermal cycler (Clever, U.K). Before PCR assay, and in order to quantify the DNA concentration (ngμl-1), the quantification of DNA samples was carried out by means of a spectrophotometric reading using 1 μl aliquots of Genomic DNA with a NanoDropTM spectrometer (NanoDrop Technologies), adopting the manufacturer’s recommendations. The concentration of DNA was estimated from absorbance at 260 nm. DNA profiles were performed using bacterial DNA and loading buffer according to the manufacturer’s instructions (Bioneer, Korea).
Detection of ompC gene by PCR: The ompC gene was detected in the present study. This gene is encoding biosynthesis of outer membrane protein C of Salmonella genus. The primer sequence of this gene (provided by Bioneer Co., S. Korea) was (ompC F: ATC GCT GAC TTA TGC AAT CG, ompC R: CGG GTT GCG TTA TAG GTC TG) 204 bp. Temperature for ompC F was 49.7ºC, and for ompC R 53.8ºC. PCR was performed at the following conditions: 95ºC 2 min 1x, 95ºC 60 s, 57ºC 60 s, 72ºC, 120 s 30x, and 72ºC 5 min 1x 8. Each primer was prepared by dissolving it in 1000 µl of deionized distilled water to obtain stocks concentration of 12 picomole/µl. Green master mix, 2x (Promega, USA) includes: 1: Taq DNA polymerase, 2: dNTPs which include 400 µM of each dATP, dGTP, dCTP, dTTP, 3: 3 mM of MgCl2 (magnesium chloride), and 4: Yellow and blue dyes as the loading dye. After PCR, the profiles of amplification products were detected by gel electrophoresis. Five µl of total reaction mixture was loaded on a 1.5% agarose gel and electrophoresed at 100 V at 70 mA for 60 min. The amplified DNA fragments were visualized by UV illumination after agarose gel electrophoresis and ethidium bromide staining by standard procedures.
Statistical analysis: Statistical analysis was carried out using SPSS version 17. Variables were presented as frequencies and percentages. Pearson’s chi square (χ2) test was applied and p-value was considered significant at p<0.01 9.
Results :
The isolation rates of Salmonella spp. were 0.5% (3/600) from human and 1% (6/600) from animals using the conventional culture methods on enrichment and selective media (Table 1).
The confirmed diagnosis of Salmonella spp. was performed by Mini API 20 E system and the results were obtained based on interpretation kit chart and results of Mini API 20 E system. The isolates showed positive results for arginine dihydrolase, lysine decarboxylase, ornithine decarboxylase, citrate utilization and H2S production. Sugar fermentations showed positive results for glucose, mannose, inositol, sorbitol, rhamnose, melibiose, arabinose, while they were negative for beta-galactosidase (ONPG), urease, tryptophan deaminase, indole production, acetoin production, and gelatin hydrolysis. They were also negative to sucrose and amygdalin.
Validity of different techniques in diagnosis of salmonella isolates: Nine isolates gave positive results for culturing and biochemical tests, Mini API 20E (at 99.9% likelihood), serotyping by polyvalent antisera (unpublished data) and gave positive results for PCR tests (Table 2).
Amplification of target DNA (ompC gene): The results of PCR amplification (performed on all DNA samples extracted from the isolates) were confirmed by electrophoresis analysis. Depending on DNA marker (8000 bp DNA ladder), the result of this estimation revealed that the amplified DNA was 204 bp for ompC gene (Figure 1).
Discussion :
In this study, it was found that Salmonella spp. infection is one of the causes of diarrhea. This may reflect the fact that Salmonella spp. is one of etiologic agents of diarrhea that infect human and animals especially during the summer, and that Salmonella is a zoonotic bacterial agent, and Salmonella typhimurium was the most common serovar found in animals and in human specimens. In this study, 3 isolates from human (0.5%) and six (1%) from animals were recovered using the conventional culture methods on enrichment and selective media. Results revealed no significant differences between human and animal isolates (p<0.01). Traditional Salmonella detection methods are based on cultures using selective media and characterization of suspicious colonies by biochemical and serological tests 7. Other studies conducted in Iraq, also revealed prevalence of Salmonella spp. at different rates of 7.9, 8.47 and 10%, 10-12 respectively. Salmonella detection by using conventional media, such as Salmonella-Shigella agar (SS), is based on lactose fermentation and H2S production and the number of false-positive results with these media that may occur necessitates conducting time-consuming and expensive additional testing 13. According to Mini API 20E results, all the 9 isolates (by culture and biochemical tests) were detected as Salmonella spp. at 99.9% likelihood level. Nucera et al 14 evaluated API 20E and PCR for the identification of Salmonella spp. isolates. They found that API 20E had the highest similarity with PCR tests at the 99.9% likelihood level and the validation of both PCR and API 20E (at the 99.9% likelihood level) showed that they are accurate diagnostic tests. Results also revealed that the isolates (No=9) were confidently identified as Salmonella spp. by detection of ompC gene. The specific PCR product (ompC gene) is a 204 bp fragment which was visualized by gel electrophoresis. All Salmonella isolates (detected by phenotypical tests) gave positive results with the PCR. The ompC gene contains sequences unique to Salmonella isolates and demonstrates that this gene is a suitable PCR target for detection of Salmonella strains.
Conclusion :
PCR technique has a high specificity in comparison with other tests, with its advantages of greater speed and effectiveness than conventional detection methods. Detection of ompC gene is a good method for detection of Salmonella species isolated from clinical specimens.
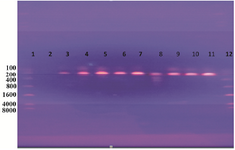
Figure 1. DNA amplification of a 204 bp of Salmonella spp. Detecting ompC gene using PCR. Lane 1: ladder; lane 2: negative; lanes 3, 4, 5, 6, 7, 8, 9, 10, and 11: positive results as Salmonella spp.; lane 12: 8000 bp marker (Ladder).
|
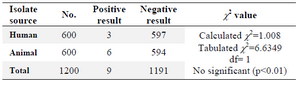
Table 1.Isolation rates of Salmonella spp. from human and animals sources
|
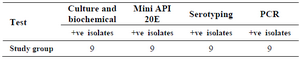
Table 2. Different lab techniques used for diagnosis of Salmonella spp
|
|