Micropatterning of ECM Proteins on Glass Substrates to Regulate Cell Attachment and Proliferation
-
G. Alamdari, Omid
-
Department of Biotechnology, College of Science, University of Tehran, Tehran, Iran
-
Department of Nanotechnology and Tissue engineering, Stem Cell Technology Research Center, Tehran, Iran
-
 Seyedjafari, Ehsan
Department of Biotechnology, College of Science, University of Tehran, Tehran, Iran, Tel: +98 21 22082120; E-mail: seyedjafari@ut.ac.ir
Seyedjafari, Ehsan
Department of Biotechnology, College of Science, University of Tehran, Tehran, Iran, Tel: +98 21 22082120; E-mail: seyedjafari@ut.ac.ir
-
Department of Biotechnology, College of Science, University of Tehran, Tehran, Iran
-
 Soleimani, Masoud
Department of Hematology, Faculty of Medical Sciences, Tarbiat Modares University, Tehran, Iran, Tel: +98 21 22082120; Email: soleim_m@modares.ac.ir
Soleimani, Masoud
Department of Hematology, Faculty of Medical Sciences, Tarbiat Modares University, Tehran, Iran, Tel: +98 21 22082120; Email: soleim_m@modares.ac.ir
-
Department of Hematology, Faculty of Medical Sciences, Tarbiat Modares University, Tehran, Iran
-
Ghaemi, Nasser
-
Department of Biotechnology, College of Science, University of Tehran, Tehran, Iran
Abstract: Background: Micropatterning is becoming a powerful tool for studying cells in vitro. This method not only uses very small amount of material but also mimic the microenvironment structure present in living tissues better than flask culturing techniques. In previous studies using micropatterning of extracellular matrix proteins on glass surfaces, the rate of protein detachment from the surface was so high that the proteins and the cultivated cells detached after 3 three days of cell seeding.
Methods: Here we optimized the glass surface modification method to fulfill the requirement of most in vitro studies.
Results: in our study we showed that the optimum time for glass surface modification reaction is 1.5 hr, and the cells could be tracked in vitro for over 15 days after cell seeding which is enough for the most in vitro studies. As a model, we cultivated HEK 293T and HepG2 cells on the collagen micropatterns and showed that they have normal growth and morphology on these micropatterns. The HEK cells also transfected with pmaxGFP plasmid vector to show that the cells on collagen micropatterns could also used in transfection studies.
Conclusion: Taking these together, this novel method is promising for efficient cell culture studies on micropatterened surfaces in the future.
Introduction :
Cells in tissues and organs are present in microenvironments which consist of Extra Cellular Matrix (ECM) and neighboring cells. These microenvironments affect different characteristics of cells including cell architecture, mechanics, polarity and function 1-6. Also, it has been shown that the arrangement and orientation of both ECMs and neighboring cells dictate cell size, cell spreading and cell morphology 7. As it is obvious, the classic cell culture cannot provide the native cell microenvironment existing naturally in different tissues. In addition, properties such as expression levels of specific genes usually come from thousands of cells. Since cells in culture vessels are usually not homogenous, these expression levels would represent cell behavior throughout the whole flask which is inaccurate.
Micropatterning of cells on surfaces have become popular since 1970s 6,8-10. The main advantage for culturing in a micro-patterned environment over growing cells in classical flask/dish culture is to establish a fine and sub-micron spatial pattern of certain molecules which leads to isolated single-cell culture, or forcing cells to grow in a certain geometric pattern 11. In comparison to culturing cells in culture flasks, micropatterning is an enabling tool for analyzing cells in their microenvironment 1 and in small scales (~500 cells) 12. Although one can control the content of the culture substratum in all scales, the small-scale analysis needs much lower amount of material and enables the researchers to perform high throughput studies 13-15. The patterned area is small so cell niches could be controlled with different materials embedded in ECM micropatterns.
Although micropatterning methods showed that the cells could be cultivated in small populations of around 500 cells/patterns, it has been shown that cell analysis could be done even in single cells 12. There are a number of methods, which have been used to make microscale patterns on different surfaces 1,6,8,16. Among them, photolithography, soft lithography and stencil printing are more used by biologists to create micropatterned surfaces. Soft lithography and stencil printing are more biocompatible than photolithography, but the patterns formed with these methods are less precise than photolithography 2,8,16.
Several groups have reported the uses of cellular micropatterns for different in vitro studies. Jones et al have studied the release of growth factors from micropatterns and its effect on alcohol injury. In their experiments, the cells were treated with alcohol to mimic alcohol injury in the presence of BMP7 and HGF, which were released from micropatterns containing collagen 17. In another experiment, Lee et al showed that the function and behavior of cultivated cells on ECM micropatterns could be analyzed through different ways such as gene expression levels 12. In their experiment, HepG2 cells were cultivated on collagen type I micropatterns and then the cells were extracted by laser catapulting for further gene expression analysis 12,18. These studies reveal micropatterning as a very strong tool for both in vitro studies and tissue engineering.
In the studies mentioned earlier, one problem was attaching the ECMs to glass surface. The main issue was that cell-ECM complexes started to detach after one day of cell seeding 12,17,18. In order to solve this problem, the glass surface treatment protocol was modified in order to minimize cell-ECM complex detachment rate. We have shown that cell behavior on collagen I micropattern spots are normal through the time points of this experiment. To investigate whether the cells on micropattern spots can go through transfection studies, they were also transfected with pmax-cGFP plasmid using lipofectamine reagent.
Materials and Methods :
Chemicals and materials: Glass slides (75×25 mm) were obtained from CitotestLabware Manufacturing Co. Ltd., (3- Acryloxypropyl) trichlorosilane was purchased from Gelest, Inc. Sulfuric acid, hydrogen peroxide, toluene, and LB medium were purchased from Merck, Dulbecco’s Modified Eagles’ Medium (DMEM) and Fetal Bovine Serum (FBS) were obtained from Gibco® Life Technologies. Liopfectamine® transfection reagent was obtained from Invitrogen. pmaxGFP was purchased from Lonza corporation.
Preparation of glass surfaces: The whole procedure for the preparation of glass surfaces and collagen micropatterning is summarized in figure 1. Glass slides were first washed with detergent, tap water, and then deionized water. The glass slides were cleaned by immersing in "piranha" solution consisting of 3:1 ratio of concentrated sulfuric acid and 35% w/v hydrogen peroxide for 10 min (Caution: Piranha solution reacts rapidly with organic material and should be handled and disposed with extreme care). The glass slides were rinsed thoroughly with deionized water, dried under nitrogen chamber. For silane modification, the slides were cleaned with an oxygen plasma chamber (Diener, Germany) at 195 W for 6 min and then immersed from 10 min to 2 hr in silane solution diluted with anhydrous toluene (20 µl of silane per 40 ml of toluene). The reaction was performed under nitrogen in a glove box (SABZ biomedicals) to prevent exposure to atmosphere’s moisture. After silane modification, the slides were rinsed with fresh toluene, dried under nitrogen and cured at 100C for 2 hr. The silane-modified glass slides were stored in a desiccator before use.
Collagen micropatterning: Collagen ECMs were dissolved in 1×PBS with 0.2 mg/ml concentration in the presence of 0.005% Tween 20. The ECMs were contact printed on glass surfaces using WhatmanMicroCaster™ Microarrayer System based on manufacturer protocol. Briefly the ECM mixture was transferred to a 96 well plate and printed using MicroCaster™ array hand tool. MicroCaster™ produces 500 µm spots, with 1000 µm pitch (spot center to center distance). After collagen printing, the slides were immersed in 1% BSA solution for 1 hr to block bare surfaces of collagen. After BSA blocking, the slides were sterilized with ethanol 70% for 20 min, washed with PBS, and kept at 4C prior to cell culture. The printed micropatterns are stable at 4C for one month.
The analysis of micropatterns attachment efficiency: Micropatterns were incubated at 37C in a humidified 5% CO2 environment for 24 hr and then stained with trichrome histology dye which specifically stains collagen ECMs. In our experiment, the percentages of undetached micropatterns were measured and considered as attachment efficiency.
Cell seeding and cultivation on collagen micropatterns: Both HepG2 and HEK293T cells were maintained in DMEM supplemented with 10% FBS, 200 units/ml penicillin, and 20 µg/ml streptomycin at 37C in a humidified 5% CO2 environment. Cells were passaged after reaching ~90% confluence. Micropatterned glass slides were placed in a 10 cm cell culture petri dish and exposed to cell suspensions of HEK 293T and HepG2 at a concentration of 1×106 cells/ml. After 1 hr of incubation, the unattached cells were removed and the slides were washed with PBS twice to remove unbound cells and placed in a new petri dish with fresh DMEM 10% FBS. In our experiments, cell growth dynamics were analyzed by counting cells on each spot for 3 days (4 Iterations). The cell counting was done by ImageJ software.
Transfection studies on micropatterned cells: pmaxGFP was cultivated in DH5α strain with LB broth medium. The plasmid was extracted using RBC plasmid miniprep kit based on the provider’s protocol. Lipofectamine transfection was done according to provider’s protocol.
Results :
Detachment rate of collagen micropatterns: Five different groups were tested for detachment rate of micropatterns. In each group, the whole process of glass surface treatment and micropatterning was the same except in silanization reaction time. Slides were stained with trichrome dye after 24 hr of incubation and the number of spots still attached to the surface was counted by light microscopy (Figure 2). According to the data (Figure 3), the amount of undetached micropatterns increased along with an increase in silanization time up to 1.5 hr and then after that the amount decreased. So the efficiency of spot attachment was highest when one hour of silanization reaction was employed (Figure 3).
Cell growth: Both HEK 293T and HepG2 cells were counted on micropattern spots at four time points (12 hr, 36 hr, 60 hr and 84 hr). Both cells showed a linear growth rate on micropatterns. HepG2 cells were more on the first day in comparison to HEK 293T cells. HEK and HepG2 cells through four time points are shown in figure 4 (A-D) and figure 4 (E-H), respectively. The number of cells on each spot is shown in figure 5 through the four time points. Both cell lines analyzed in this experiment exhibited normal morphology.
Transfection capabilities of micropatterned cells: Both HEK 293T and HepG2 cells were transfected using pmaxGFP vector and lipofectamine. The cells remained normal in morphology after transfection. The HEK 293T transfection rate after 24 hr was higher than 90% (Figure 6). Percentage of the cells expressing GFP protein 48 hr after transfection was near 99% (data not shown).
Discussion :
Micropatterning usefulness appears when it comes to mimicking the microenvironment present in the tissues 1,7,8,16. As mentioned earlier, preparing this microenvironment helps the in vitro studies to elucidate what happens in living organisms. Microenvironment in different tissues dictates the fate of the cells by its chemical and physical characteristics. These characteristics dictate cell division rates and cell spreading behaviors. The behavior of tissue in growth pattern and hormone diffusion depends on these characteristics as well. In a study discussed earlier 17 Jones et al used collagen micropatterns embedded with BMP7 and HGF growth factors to mimic hepatic microenvironments for hepatic differentiation. In other studies, micropatterning is used as a tool for high throughput studies including cell cloning 13, gene transfection 15, cell secretion studies 14 and tissue engineering 19.
In previous studies using ECM micropatterning, one of the main problems was that the cells could not be tracked after three days due to ECM micropattern detachment which became a trouble when it came to in vitro differentiation studies. In our method, the cells were attached to surface for 15 days after cell seeding, which made it possible to track cells for transfection, differentiation, and gene expression studies that require cell tracking of 10 days or longer. In this study, we showed that by extending the surface modification reaction (silanzation) to 1.5 hr, the detachment rate of micropatterned ECMs would be near to zero (Figure 3).
Although the process of glass surface treatment and micropatterning is time consuming and laborious, its advantage, that the cells could be tracked using optical microscopy, makes it valuable in comparison to techniques using metal coated glass surfaces (e.g. gold) 20. The whole process of glass surface treatment and attaching the ECM proteins through lysine and cysteine groups are shown in figure 1. Although the number of HEK and HepG2 cells seeded on collagen I spots were the same, after 12 hr, HepG2 cells were more on the spots. This shows the higher tendency of HepG2 cells in attaching to micropattern ECMs.
This differential tendency of cells for different ECMs can be used for micropatterned co-culture studies. As an example, Lee et al 19 used a printed pattern of collagen I and fibronectin. They cultivated mESCs (mouse Embryonic Stem Cells) and HepG2 cells sequentially on these spots (mESCs were seeded first followed by washing unattached cells, then they seeded HepG2 cells followed by another step of washing unattached cells). Since mESCs have higher tendencies for fibronectin, they would attach only to fibronectin (washed with appropriate timing) and HepG2 will only attach to collagen I micropatterns (due to higher tendency). Later, they showed that this co-culture could facilitate the differentiation of mESC cells to hepatic cells by analyzing hepatic gene expression levels.
The model in transfection studies which uses pmaxGFP vector reveals that cells could go through transfection studies while culturing on micropatterns. The transfection occurred with more than 90% of efficiency in HEK cells, which again makes the method valuable since transfection of cells located on micropatterns consumes low amount of both lipofectamine and plasmidic DNA in comparison to other techniques involved in ECM and non-viral gene 21.
Conclusion :
In the present study, the glass surface modification method was optimized to fulfill the requirement of most in-vitro studies in the field of micropatterning. Taking all the results into account, this novel method is promising for efficient cell culture studies on micropatterened surfaces in the future.
Acknowledgement :
The authors would like to acknowledge the financial support of University of Tehran and Stem Cell Technology Research Center for this research.
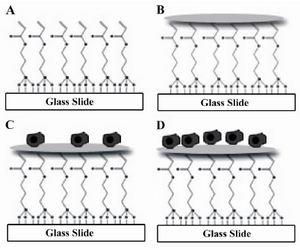
Figure 1. Glass surface modification and cell seeding process. A) Glass surface modification; B) Collagen ECM micro-patterning; C) Cell seeding; D) Cell spreading and division
|
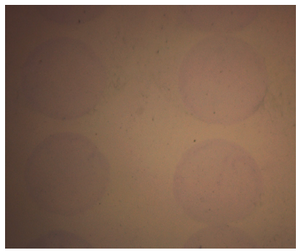
Figure 2. Collagen micropatterns stained with trichrome
|
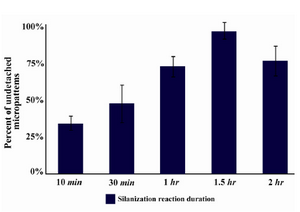
Figure 3. Percentage of spots detached from glass surface after 24 hr of incubation DMEM 10% FBS
|
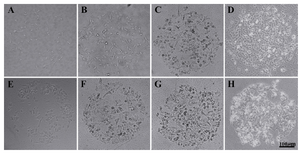
Figure 4. HEK 293T through four time points, A) 12 hr; B) 36 hr; C) 60 hr; and D) 84 hr; HepG2 cells through four time points, E) 12 hr; F) 36 hr; G) 60 hr; and (H) 84 hr
|
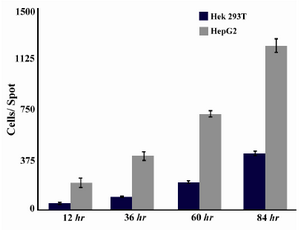
Figure 5. Cell growth behavior in HEK 293T and HepG2 cell lines measured by ImageJ software
|
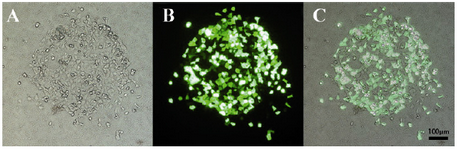
Figure 6. HEK 293T transfection with pmaxGFP plasmid; A) Bright field; B) Fluorescent field; C) Image overlay
|
|