Extracellular L-Asparaginase Synthesis Bacillus niacin Isolation, Optimization, and Characterization from Marine Saltern Sediment Sources
-
 Palaniyandi , Thirunavukkarasu
Department of Biotechnology, Dr. M.G.R Educational and Research Institute, Chennai, Tamil Nadu, India, Tel: +91 44 8610749156; E-mail: thirunavukkarasu.ibt@ drmgrdu.ac.in
Palaniyandi , Thirunavukkarasu
Department of Biotechnology, Dr. M.G.R Educational and Research Institute, Chennai, Tamil Nadu, India, Tel: +91 44 8610749156; E-mail: thirunavukkarasu.ibt@ drmgrdu.ac.in
-
Department of Anatomy, Biomedical Research Unit and Laboratory Animal Centre, Saveetha Dental College and Hospital, Saveetha Institute of Medical and Technical Science, Saveetha University, Chennai, Tamil Nadu, India
-
Wyson , John
-
Department of Food Processing Technology, AMET University, Kanathur, Chennai, Tamil Nadu, India
-
Sivaji, Asha
-
Department of Biochemistry, DKM College for Women, Vellore-632001, Tamil Nadu, India
-
Thamada, Swarnakala
-
Molecular Systematics Laboratory, Zoological Survey of India, Andaman & Nicobar Regional Centre, Port Blair - 744 102, Andaman and Nicobar Islands
Abstract: Background: Asparagine is an amino acid that can be converted into aspartic acid and ammonia by the enzyme L-asparaginase. Some forms of cancer, such Acute Lymphoblastic Leukaemia (ALL) and Non-Hodgkin Lymphoma (NHL), respond well to this enzyme when employed as a chemotherapeutic drug. The purpose of this research was to find bacteria that can manufacture the enzymes L-asparaginasein marine slattern sediment which can be employed in commercial and industrial scale production.
Methods: All of the strains were identified as Bacillus niacini spp. by biochemical and molecular testing. The strain belongs to the Bacillus genus, according to nutritional, biochemical, PCR and 16srRNA sequencing data.
Results: According to the findings of this research, Bacillus niacin spp. have the potential to create a substance that is helpful in a variety of medical applications. The results of this study hint to the possibility that bacteria have the ability to produce antimicrobial compounds, which have the potential to be successful in a wide variety of environments.
Conclusion: Numerous opportunities may arise for researchers interested in utilizing the medical potential of enzyme-producing bacteria if they are successfully isolated and screened from aquatic and terrestrial habitats.
Introduction :
L-asparaginase hydrolyzes L-asparagine to L-aspartic acid and ammonia. This process produces ammonia, which can be difficult if not fully monitored and treated during therapy. High blood ammonia can cause confusion, sleepiness, and coma. L-asparaginase therapy requires ammonia monitoring and toxicity management. L-asparaginase with lower ammonia levels may reduce this side effect. In recent years, a lot of attention has been paid to its potential to prevent cancer due to its anti-carcinogenic properties 1. Lymphoid cancer cells, like all tumour cells, lack the ability to synthesize L-glutamic acid and therefore rely on external sources of glutamine for their rapid proliferation. Consequently, the activity of L-glutamine is to cause the death of tumour cells that rely on L-glutamine for survival 2. Because of their strong substrate affinity, micro-organisms Escherichia coli (E. coli) and Erwinia spp. are used as drugs for lymphoblastic leukaemia 3.
L-asparaginase is present in the following: a variety of animal tissues, micro-organisms, plants, and rodent serum, but not in humans. Enterobacter aerogens, Pseudomonas stutzeri, Staphylococcus aureus, and Serratia marscescens are just a few of the microbes that have been shown to produce L-asparaginase 4. Since microbial enzymes are very crucial, this investigation has concentrated on isolating and testing bacteria from both land and water that generate L-asparaginase, as well as their antibacterial capabilities. Until devised for patients, however, no one was ever written down to have recovered 5. Acute Lymphoblastic Leukaemia (ALL) is one type of human malignancy that has benefited greatly from the use of microbial L-asparaginase as a therapeutic agent 6. This is because it readily biodegrades, poses no health risks to humans or animals, and may be administered with no effort on the spot. Bio-active chemicals with commercial value may be produced by marine bacteria 7. As the most prolific source of antibiotics and other bio-active secondary metabolites, bacteria have risen to prominence among marine microorganisms. But whereas many bacterial studies have focused on antibiotic synthesis, relatively few investigations have explored bacteria's enzymatic potential 8. L-asparaginase, an enzyme produced by Proteus vulgaris, has enzyme properties. Microbes such Pseudomonas aeruginosa, E. coli, Citrobacter spp. and Bacillus spp. were examined for enzyme activity. Despite the fact that L-asparaginase activity has been discovered in plants, animals, and micro-organisms (bacteria, fungi, and actinomycetes), and even in the serum of certain rodents, asparaginase has not yet been isolated from a human source 9. Although asparaginase synthesis by bacteria isolated from marine sediments has been studied previously, no such research has been published on seawater-derived micro-organisms that produce enzymes. Isolating and screening marine micro-organisms to produce anti-cancer L-asparaginase from seawater is the subject of the current study 10.
Materials and Methods :
Collection of samples: Twenty samples of coastal sediment were gathered from Marakknam slattern in Tamil Nadu, India (at 12.1899 degrees N and 79.9249 degrees E). The samples were gathered in a controlled environment and shipped off to the lab in sealed, sterile containers.
Obtaining samples: At a depth of 10 cm, twenty marine sediment samples were obtained from the coastal region of Marakkanam slatterns, (12.1899 N, 79.9249° E) Tamil Nadu, India.
Preparing the media, medium for the production of asparaginase (m-9), isolation of bacteria: The samples were serially diluted and then plated on starch casein agar, glucose-asparagine agar, and glycine-glycerol agar to identify the bacterial species responsible for producing L-asparaginase. Next, the samples were kept in an incubator at 37°C for seven days. After incubation, bacterial colonies were observed on the agar plates. The colonies were picked and sub-cultured on fresh agar plates to obtain pure cultures. The bacterial isolates were then screened for L-asparaginase activity using the plate method 11.
Secondary screening of micro-organisms that produce L-asparaginase: L-asparaginase production medium was made again and placed into plates, each with two colonies streaking. The plates were incubated for another two days. The pure pink ring surrounding the colony indicated that L-asparaginase enzymes were being synthesized 12.
L-asparaginase fermentation: M9 minimal media (M9) is used to cultivate bacteria. Amino acids and other carbon sources may be added to the M9 minimal salts base. The components of the new M9 medium are as follows. The broth was injected with the selected bacterial isolate 19 and incubated for three days, and centrifuged at 5000 rpm for 10 min after witnessing growth in broth medium 13.
Enzyme assay for asparaginase: Wriston and Yellin used nesslerization to assess the quantity of freed ammonia to determine extracellular L-asparaginase activity 1.5 ml of 0.04 M L-asparagine produced in 0.05 M Tris-HCl buffer, pH=8.6, and 0.5 ml of the enzyme to make up a total volume of 2 ml. For 30 min, the tubes were incubated at 37°C. The reaction terminated using 0.5 ml of 1.5 M Trichloroacetic Acid (TCA). The blank was run by adding TCA followed by enzyme preparation. The reaction contents were centrifuged at 10,000 g for 5 min to remove the precipitated protein, and the filtrate was collected. 0.5 ml filtrate was diluted to 7 ml with distilled water for detection of freed ammonia, and 1 ml Nessler's reagent was added to the resultant mixture. The colour reaction was allowed to run for 20 min before the absorbance at 480 nm was measured with an Optizen Pop -UV/Vis spectrophotometer. The presence of ammonia is indicated by a yellow tint. At larger concentrations, however, a brown precipitate forms. The amount of ammonia liberated by the test sample was estimated by comparing the absorbance to a standard curve constructed from ammonium chloride solutions as the ammonia source. One unit (U) of L-asparaginase is the quantity of enzyme required to produce one mole of ammonia in one minute at 37°C and pH=8.6 14.
Manufacturing of L-asparaginase has been optimized: M9 medium was created and sterilized with several substrates such as starch, fructose, glycerol, and sucrose. The chosen organisms were injected into the medium and maintained for three days in a shaker (120 rpm) at room temperature. After 3 days, the sample was drawn aseptically and the enzymes were measured using a UV-Visible spectrophotometer. The influence of medium pH on the development and synthesis of L-asparaginase enzymes has been demonstrated 15.
Optimizing nitrogen for L- asparaginase: Different nitrogen substrates, such as yeast extract, gelatin, ammonium sulphate, and potassium nitrate, were produced and sterilized in modified M9 medium. The chosen organisms were then injected into the medium and stored in a shaker for three days at room temperature (120 rpm). After three days, the sample was taken aseptically and L-asparaginase was measured using a UV-Vis spectrophotometer. Effect of incubation period in Erlenmeyer flask (200 ml) with production medium (50 ml) was incubated in a shaker incubator (70 rpm) at 37°C for 3 days to find the best incubation time for making the most L-asparaginase 16.
Optimizing production conditions: The bacterial isolate's ideal incubation temperature was used, and 0.5 ml of the inoculums was introduced to 5 ml of sterile basal medium that had been optimally supplemented with various carbon and nitrogen sources 17.
L-asparaginase partial purification: A four-day-old culture filtrate (200 ml) cultivated in L-asparagines-containing M9 medium broth was recovered by centrifugation at 8000 rpm for 10 min at 4°C. The crude enzyme was precipitated with ammonium sulphate, and the protein precipitate was allowed to stand overnight at 80% salt saturation. The precipitate was re suspended in 0.01 M Tris-Hcl buffer (pH=7.2) and dialyzed overnight against the same buffer after centrifugation at 10,000 rpm for 15 min 18.
Dialysis for a protein sample, a typical dialysis method: Wet the membranes with phosphate buffer beforehand (20 mM, pH=6.5). Fill the dialysis tubes or devices with the sample. Dialyze with phosphate buffer for 2 hr at room temperature (20 mM, pH=6.5). Finally, replace the buffer and dialyze at 4°C overnight. The dialysis bag sample was then used as a protein source for future research 19.
Protein estimation by Bradford: Bradford dye binding assays measure protein quantities. Acidic conditions turn protein-binding CBB dye blue. Bradford reagent and Bovine Serum Albumin (BSA) create a standard curve. Proteins increase the dye's blue colour. The Bradford reagent measures blue hue intensity at 480 nm to assess a material's makeup. Standard curves compare protein concentration to blue colour intensity. A spectrophotometer showed blue at 480 nm after combining 1 m l culture filtrate with 5 ml Bradford's reagent. Comparing blue colour intensity to the BSA standard curve determined protein content 18.
SDS-polyacrylamide gel electrophoresis technique: Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) is a widely used technique for separating and analyzing proteins based on their molecular weight. Proteins are denatured and treated with SDS, a detergent, which unfolds and imparts a negative charge to the proteins, making them linear and negatively charged. A reducing agent, such as β-mercaptoethanol, is often used to break disulfide bonds in proteins, further denaturing them. A polyacrylamide gel is created by polymerizing acrylamide and bisacrylamide in the presence of a catalyst. The concentration of acrylamide in the gel determines the separation range for proteins. Higher concentrations are used for resolving smaller proteins. Samples are loaded into wells in the gel, typically alongside a protein ladder with known molecular weights for reference. An electric field is applied across the gel. SDS-covered proteins migrate through the gel according to their size, with smaller proteins moving faster than larger ones. After electrophoresis, the gel is stained with a dye, such as Coomassie Blue stain, which binds to proteins and allows them to be visualized. The separated protein bands are compared to the molecular weight standards, allowing estimation of the molecular weight of the sample proteins 20.
Biochemical and physiology characterized of selected isolation 19th sample: This test reveals if bacteria can use citrate as their sole carbon source. Green-to-blue mediums imply success. Methyl red test: Bacteria digest glucose into acid. Red medium means success. Cytochrome oxidase is detected by this test. Blue-purple medium denotes success. Voges Proskaure test (VP): Measures glucose fermentation-produced acetoin. Reddish middle symbolizes success. Urease test: Urease converts urea into ammonia and carbon dioxide. Pink medium means success. Tryptophan-to-indole test: Bacteria make indole. Positive findings turn medium red with Kovac's reagent. Gas is produced when bacteria ferment glucose, lactose, and sucrose. Gas, pH, and colour determine test results. Biochemical and physiological features of the bacteria sample can be determined and classified. A positive oxidase test and a negative citrate utilization test may indicate the bacteria 21.
Isolation of bacterial DNA: DNA isolation is a physical and chemical method of purification. The ethanol precipitation method was used to isolate bacterial DNA from L-asparaginase enzyme generating bacteria. Collect the loop containing the 24 hr bacterial organism and dissolve it in 600 µl of 50 mM NaOH in a centrifuge tube. Place the bacterial culture in a 94°C water bath for 3 to 5 min. For 5 min, keep the lysate bacterial culture at -20°C. After adding 32 µl of TrisHCl, centrifuge at 2000 rpm for 30 min. Collect the aqueous solution and then add 70% ethanol to it. After five minutes of centrifugation at 5,000 rpm, the pellet is collected. TAE buffer was used to dissolve this DNA. Loading the agarose gel with 8 µl of DNA sample and 2 µl of loading dye (Bromophenol blue), the agarose gel (0.8%) was stored in an electrophoresis apparatus with tank buffer (1× TAE buffer) and operated at 100 V. The agarose gel was placed under the UV-light spectrophotometer 22.
Using the 16s Ribosomal RNA gene, Bacterial isolates can be identified: The PCR amplification was carried out with a reaction mixture that was 20 µl in volume and contained 4 µl of template DNA, Nuclease Free 7 µl, MgCl2 (25 mM) 2.8 µl, 0.8 µl of 16S r RNA primers, 3. 2 µl of deoxynucleoside triphosphates, 1.5 mM of MgCl2, 0.2 µl of Taq DNA polymerase (MBI Ferments), and 2 µl of 10× Taq polymerase buffer. The following is the sequence of the 16S rRNA primers that were utilized: 27f: (5'-AGAGTTTGATCCTGGCTCAG-3')1522r: (5′-AAGGAGGTGATCCANCCRCA-3').
An initial denaturation at 95°C for five min was used in the amplification process. This was followed by 35 cycles of denaturation at 94°C for 45 s, anneal ing at 56°C for 45 s, extension at 72°C for one min, and a final extension at 72°C for five min using a thermo cycler (iCycler; Bio-Rad Laboratories, CA). PCR results were examined for 16S rRNA amplicons on an agarose gel using 1% agarose and 1×TAE buffer at 100 V 23.
Production of PCR sequence analysis: Using the QIA fast gel extraction kit (QIagen, Valencia, CA), the 16S rRNA and ITS region amplified fragments were purified from the agarose gel and sequenced using an automated DNA sequence (Model 3790, Applied Bio system, USA). The sequences were analysed with the help of the BLAST software, which can be found on the NCBI website (http://www.ncbi. nlm.nih.gov/blast). This software is an option for the basic Local Alignment Search Tool 24.
Genetic analysis of evolutionary relationships: In order to align the sequences of these 16S rRNA genes, the BLASTN programme 25 and the CLUSTAL W software 26 were used. The BLASTN programme was used to match the sequences of these genes to those that are accessible from GenBank. Distances were determined using Kimura's two-parameter correction 27, which was applied to the calculations. The neighbor-joining approach was utilized when it came to the construction of phylogenetic trees 28. The bootstrap analysis was carried out using a total of one thousand replications. For each and every analysis, the MEGA4 programme was utilized 29.
Results :
Sample collection: At a depth of 10 cm, the samples were taken from Marakkanam, which is located at 12.1899 degrees north latitude and 79.9831 degrees east longitude in Tamil Nadu, India (Figure 1).
Secondary screening of L-asparaginase producing bacteria: Screening results for L-asparaginase production were significantly higher in 19 of the 20 identified strains (Figure 2). Secondary screening was performed on the same enrichment agar media to ensure that the isolate's capacity to produce L-asparaginase was consistent. Isolate 19 was chosen for further study because it displayed notable activity in both of the secondary screenings for L-asparaginase production (Figure 3).
Optimization of L-asparaginase enzyme: Carbon source: Among the carbon sources tested the original carbon source of the medium M9 starch found to be best source which yielded 0.115 U/ml L-asparaginase activity, followed by galactose which showed 0.068 U/ml. The least L-asparaginase activity was observed on maltose as 0.061 U/ml, (Figure 4, Table 1).
Effect of pH source: Culturing the strain in M9 broth with varied pH values ranging from 5.0 to 8.0 allowed researchers to evaluate the effect of pH on the synthesis of L-asparaginase. This step's ideal pH was utilized as the basis for further research (Table 2).
Effect of nitrogen source: The lowest L-asparaginase activity was found at a concentration of 4 g yeast extraction and the greatest was found at a concentration of 6 g/L potassium (3.70 U/ml). The research showed that ionic nitrate is necessary for the growth of bacteria to produce L- Asparaginase (Table 3).
Effect of incubation temperature: The organism and temperature both affect the activity of L-asparaginases, with the highest temperature having the least effect on enzyme activity. The L-asparaginase activity was at its best at a temperature of 35°C, when it registered 3.78 U/ml (Table 4).
Producing at optimum conditions: Bacillus niacin spp. was reported to have the maximum L-asparaginase activity of 4.22 U/ml when the conditions were at their best. In order to produce the most L-asparaginase, the bacterial isolates thrive best in medium that is slightly alkaline to alkaline (pH7) (Table 5).
Electrophoresis with SDS-polyacrylamide gel: SDS-PAGE analysis revealed that the enzyme exists as a single band that possesses electrophoretic mobility. The apparent molecular weight of Bacillus niacini L-asparaginase was determined to be 56.5 kDa by using a variety of reference proteins whose molecular weights were already known. This was discovered through the use of these proteins (Figure 5).
Biochemical and physiological characterization: The following biochemical tests were carried out: Citrate utilization test, Methyl red test, Oxidase test, Voges Proskaure test (VP), Urease test, Indole test and Triple Sugar Test (TSI) (Table 6, Figure 6).
Isolation of bacteria DNA: The bacterial DNA from production of bacteria was carried out by ethanol precipitation method. High molecular weight genomic DNA was isolated using the alkali lysis method (Figure 7).
PCR Amplification of 16S rRNA gene: PCR amplification was performed using a 50 µl reaction mixture and agarose gel electrophoresis showed presence of amplified product of 16s rRNA gene with the molecular weight of approximately 1.5 KB (Figure 8).
Conditions: 1.5% agarose gel electrophoresis
(Lane a: 100 bp DNA Ladder; b: Sample)
1 KB DNA Ladder (bp):2000, 1500, 1000, 900, 800, 700, 600, 500, 400, 300, 200, 100.
Sequence analysis of PCR products: To do this, we used NCBI's Basic Local Alignment Search Tool (BLAST) to compare the sequences (http://www.ncbi.nlm.nih.gov/blast).
Sequences of PCR production
>Isolate 19th Sequences
>M4_16SF_S10339
>NR_113777.1 Bacillus niacini
Utilizing the Neighbour-Joining approach allowed the inference of the evolutionary history. This illustration depicts the best possible tree, which has a total branch length of 0.18542627. Next to each node in the tree, you'll see the percentage of times that a similar set of trees (1000 replicates) produced the same clustering of linked taxa. This proportion could be ranging from 0 to 100% of the total. The phylogenetic tree has been drawn to scale, with branch lengths shown using the same units as the evolutionary distances that were used to estimate the phylogenetic tree and this allows for a more accurate representation of the tree. The Kimura 2-parameter approach was used to calculate the evolutionary distances, and the results are expressed in terms of the number of base substitutions that occurred at each location. Included codon positions were the first, second, third, and noncoding locations. Only in pair wise sequencing comparisons were all places that have alignment gaps and missing data removed (Pairwise deletion option). The completed dataset contained a total of 1486 locations in its entirety. The programme MEGA4 was used to run the phylogenetic analysis.
The BLAST analysis that was performed on the NCBI database, as well as the RDB taxonomy analysis and the phylogenetic tree, unequivocally demonstrated that the sample in question belonged to the taxon of Bacillus niacin (Figure 9).
Discussion :
Choosing an appropriate niche that is likely to create potential producers of any metabolite is one of the most effective strategies for determining acceptable candidates for any target metabolite. This is because this method is one of the few that actually works. Numerous investigations on asparaginase in soil have been carried out. The purpose of this research was to identify bacterial isolates with the potential for commercial and industrial production of L-asparaginase enzymes. During this study, 20 different L-asparaginase-producing bacteria were identified from maritime sediment samples collected in Tamil Nadu. Oceanic bacteria was the source of one of the L-asparaginase-producing isolates. This is owing to the fact that sea silt provides a favorable environment for nutrient-rich microorganisms. All of the strains were identified as Bacillus spp. by biochemical and molecular testing (Deshpande N, Choubey P, Agashe M., 2014), who identified Bacillus spp. with L-asparaginase activity, found similar results. In addition, this research looks into 33 other topics. A marine bacterial strain was isolated and morphologically characterized. When the strain's shape and spores were examined under a light microscope, they showed normal bacillus morphology, despite the fact that bacillus is a non-spore producing bacteria. The nutritional, biochemical, and 16srRNA sequencing results indicate that the strain belongs to the bacillus genus. Bacillus was confirmed and identified as the strain. Bacillus spp was tested for L-Asparaginase production using both plate and submerged fermentation methods. Similarly, several bacterial genera such as Bacillus circulans and Streptomyces sp. have been shown to produce L-Asparaginase. Isolates from the sea when compared to other organisms, L-Asparaginase activity is higher. L-Asparaginase was produced via submerged fermentation, yielding crude enzyme with a total activity of 11.20 mol ammonia/ml.
Conclusion :
On basis of the study that has been presented here, the following conclusions have been formed. The vast majority of research on L-asparaginase comes from bacterial sources; however, plant sources have also been proven to be good sources for the isolation of L-asparaginase. Since bacterial L-asparaginase is associated with a high number of adverse effects, the purified enzyme will make an excellent therapeutic treatment. After 120 hr of incubation, the L-asparaginase activity reached its peak level (11.202 mol ammonia/ml), as determined by the findings of the current investigation, which led us to the following conclusion.
In the end, we came to the conclusion that the powerful isolate is a member of the genus Bacillus, species Salivarius, and the Genbank accession number that was given to it. It may be possible to gain a comprehensive understanding of the therapeutic potential of enzyme-producing bacteria by isolating and screening them in their natural environments (both aquatic and terrestrial). The findings of this study point to the bacteria's potential ability to create antimicrobial chemicals, which have the potential to be effective in a wide variety of medicinal applications.
Acknowledgement :
We great fully acknowledge Er. A. C. S. Arun Kumar, President, Dr. M. G. R. Educational and Research Institute, University for providing the necessary facilities.
Funding: The authors declare that no funds, grant, or other support were received during the preparation.
Conflict of Interest :
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.

Figure 1. The samples were taken at a depth of 10 cm from Marakkanam (12.1899°N 79.9831°E), Tamil Nadu, and India.
|
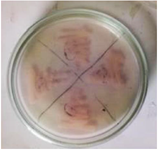
Figure 2. L-asparaginase secondary screening of the isolate plate.
|
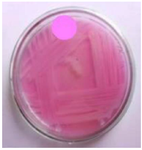
Figure 3. 19th Isolate were selected on secondary screening medium for L-Asparaginase activity.
|

Figure 4. Carbon source's influence on L-asparaginase activity in the 19thculture.
|
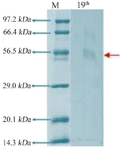
Figure 5. Electrophoresis with SDS polyacrylamide gel 19th culture.
|

Figure 6. Biochemical and physiological characterization of the selected isolate 19th culture.
|
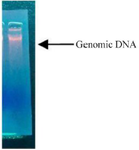
Figure 7. Genomic DNA of given bacteria isolate 19th culture.
|

Figure 8. PCR amplification profile of given bacteria isolate 19th culture.
|

Figure 9. Phylogeny tree analysis for 19th culture.
|
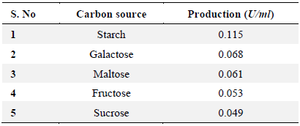
Table 1. Carbon source affects 19th culture L-asparaginase synthesis
|
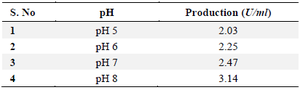
Table 2. Effect of pH on L-asparaginase production for selected 19th culture
|
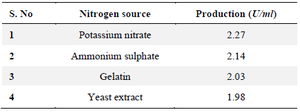
Table 3. Effect of concentration on nitrogen sources for selected 19th culture
|
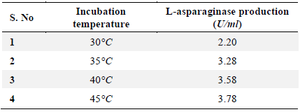
Table 4. Effect of incubation on L-asparaginase production for selected 19th culture
|

Table 5. Production at optimum condition for selected 19th culture
|
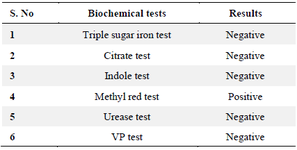
Table 6. Biochemical and physiological characterization of selected 19th isolate
|
|